Review Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
How Do We Clean Up This Mess? - A Review of The Testing Methodologies Used for Detection of Live Bacteria in Healthcare Environments
*Corresponding author: Andrew Kempa, Q Technologies group, University of Lincoln, Lincoln, UK.
Received: July 23, 2019; Published: August 06, 2019
DOI: 10.34297/AJBSR.2019.04.000808
Abstract
In light of the increasing numbers of disinfectant resistant bacterial species, and the proven direct link between disinfectant resistance and antibiotic resistance, the detection of live bacteria in high risk environments within healthcare, is of increasing importance. The benefits of accurate identification, and in some cases quantification, is paramount in indicating the effectiveness of cleaning products, protocols and therefore safety to practice for patients and staff. A review of the currently available environmental testing methods, highlighting those tests most appropriate for the different areas of our healthcare services has been explored. Looking at the latest technological advances in surface and microbiological testing, to determine their efficiency in adequately detecting organisms of potential significance, and therefore informing us of the most appropriate ways in which to treat areas that have been potentially contaminated. The outcome of using the correct testing methods can determine the true impact of surface contamination in primary and cross infection. Moreover, informing us of the effectiveness of cleaning interventions in the hope of decreasing potential causes of infection (including staff behaviours) and subsequently improving patient outcomes.
Keywords: Disinfectant resistance; Bacterial detection; live bacteria; hospital environments; surface detection
Historical Perspective
We have been aware of microorganisms and their impact on human health for over 150 years. Anton Van Leeuwenhoek was the first person to document microscopic observations of bacteria in 1676 (although a Jesuit priest, Athanasius Kircher is also credited by some as the first to observe microorganisms at around the same time). In 1878 Robert Koch discovered how to grow bacteria in a Petri dish (named after his assistant Julius Petri). Circa 1900 Martinus Beijernick extended the work of both Koch and Pasteur to develop what is now the enrichment culture techniques used today. However, in the present day, it is generally believed that the vast majority of microorganisms present in common environments cannot be successfully cultured and characterised using current standard testing methods [1-5]. Ever since the discoveries of Semmelweis in 1847, and in spite of our industrial, academic and clinical efforts to reduce HAI’s (Hospital Acquired Infections), we have had varying degrees of success. There have been longstanding theories and hypotheses surrounding the contributing factors to HAIs; for example, effectiveness of hand hygiene and the impact of airborne contaminates amongst many others. Although there has been a significant amount of research done in the food preparation sector, until recently there has been limited evidence focused on surface bio burden, and its relevance in specialised critical areas of healthcare [6-8].[6,8].
The root cause of this is multifactorial, although one main reason can be associated with the technological advances, or lack thereof in the standard testing methodology used to confirm successful cleaning of clinical areas. In 2004, when Dancer proposed a standard for microbiological assessment of surfaces, many routine molecular techniques were not available outside either academic research or the military [5]. It took over a decade more, before the creation of the UK Environmental Special Interest Group (ESIG) with a remit to explore further advances and changes around updated standards (Data unpublished). It is therefore unsurprising that many healthcare facilities and researchers still consider culture as the “gold standard” for confirmation of the absence of potential pathogens that could cause HAIs [9]. These test methodologies are used predominantly to initially detect the presence or absence of microorganisms and to subsequently identify ideally to a species level. However, techniques such as culture have not changed significantly in many years and continue to have significant limitations in both sensitivity and specificity. Although a pure bacterial culture remains important for the study of virulence, antibiotic susceptibility, and genome profiling in order to facilitate the understanding and treatment of caused diseases, there continues to be little correlation between in-vitro evidence and its relevance in-situ.
The recent availability of molecular diagnostic technology has provided greater sensitivity and specificity in the detection of microorganisms. Evidence would suggest improvement in detection methods provides greater insight into the efficiency of current cleaning and decontamination of clinical environments [10]. Improved detection methods may also help us to understand the true extent of bacterial resistance to disinfectants. It is now considered “routine” for molecular diagnostics to deliver results in hours and minutes compared to traditional culture results taking days and in some cases weeks.
Current Available Tests
Visual inspection
Unfortunately, due to many factors including the lack of a fast, accurate test, visual inspection is still used by the vast majority of healthcare institutions, even though the evidence is clear that it is not an accurate test of cleanliness, let alone bioburden [11,12].
Total ATP
Adenosine Tri Phosphate is present in all living cells except viruses. It is measured by the inexpensive test kits, in relative light units (RLU), and is a fast, simple way to measure extra cellular and some intra cellular ATP. Total ATP testing has been undertaken for more than a decade in the food industry, yet no statistically relevant direct relationship has been found between RLU and Colony Forming Units (CFU) [13,14]. As there is no certainty that the test equipment is able to measure intra cellular ATP, even a zero count cannot be considered relevant.
Culture
The first culture conditions used various parameters such as incubation time, nutrients, atmosphere, and temperature. Refinement around such parameters has continued since its conception over 100 years ago. The use of culture in clinical microbiology was prompted by microbiologists initially specializing in intracellular bacteria [15,16]. The shell vial procedure allowed the culture of new species of Rickettsia. The design of axenic media for growing fastidious bacteria such as Tropheryma whipplei and Coxiella burnetii, and the ability of amoebal coculture to discover new bacteria constituted major advances. Strong efforts associating optimized culture media, detection methods, and a microaerophilic atmosphere allowed a dramatic decrease of the time of Mycobacterium tuberculosis culture. The use of a new versatile medium allowed an extension of the repertoire of archaea. Finally, to optimize the culture of anaerobes in routine bacteriology laboratories, the addition of antioxidants in culture media under an aerobic atmosphere, allowed the growth of strictly anaerobic species [17-19,]. There is increasing need to improve the relevance of in-vitro testing and provide much greater correlation between the evidence gained within the laboratory environment, and that of the clinical setting.
This should ultimately lead to a better understanding of the colonisation of clinical surfaces, and how those organisms change in characteristics and concentration over time. Although there are a large number of reasons that surface bacterium may not be easily cultured in the lab, most of these can be reduced to the difficulty of replicating the very precise environment conditions certain microbes require for growth. Early culturing efforts focused on microbes that were easily grown under standard conditions, largely due to adaptation to the incubator-like conditions of an animal body. By contrast, free-living environmental microbes can have a broad array of environmental requirements. One example of interest is the discovery that many organisms cannot survive in the high-nutrient conditions favoured by standard cultural practices [1,2,20,21]. The development of low-nutrient media has greatly increased the number of organisms that have been successfully cultured. Many soil organisms are incapable of surviving under high-oxygen conditions. These obligate anaerobes would naturally not survive standard culturing approaches, but there have been some successes with growing them in anaerobic chambers [22]. However, in respect to cultures usefulness in accurately testing the efficacy of disinfectants, the US CDC states, “Attempts to substantiate the bactericidal label claims of phenolics using the AOAC Use-Dilution Method occasionally have failed” [23]. However, results from these same studies have varied dramatically among laboratories testing identical products (Figure 1 ) [24].
Polymerase Chain Reaction (PCR)
PCR is a technique used to amplify a segment of DNA across several orders of magnitude [25]. This then generates thousands to millions of copies of a particular DNA sequence. Developed in 1983 by the Cetus Corporation [26], it is a reliable way to repeatedly replicate a focused segment of DNA. This technique is used in biomedical research, criminal forensics, and molecular archaeology [27]. PCR is now a common and often indispensable technique used in clinical and research laboratories for a broad variety of applications. For example, these applications can include DNA cloning for sequencing, gene cloning and manipulation, functional analysis of genes and detection and characterisation of microorganisms causing disease. The vast majority of PCR methods rely on thermal cycling, which involves exposing the reactants to cycles of repeated heating and cooling, permitting different temperature- dependent reactions, specifically, DNA melting and enzyme-driven DNA replication [28]. Short DNA fragments containing sequences comple mentary to the target region, along with a DNA polymerase, enable selective and repeated amplification. As the PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the original DNA template is exponentially amplified.
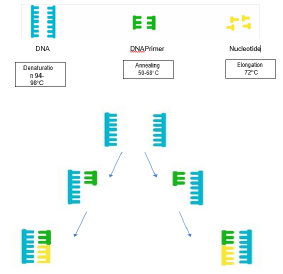
Figure 2: Conventional PCR Cycle, involving DNA, primers and nucleotides, with typical temperatures associated with each reaction step.
The simplicity of the basic principle underlying PCR means it can be extensively modified to perform a wide array of genetic manipulations previously described. In terms of detection and characterisation of organisms causing disease, PCR has been used in environmental sampling for some time and in certain situations has been an advantage when compared with more traditional culture techniques. Such examples would be detection of legionella from water sources, where often culture techniques would take several days, whereas a PCR can take several hours (Figure 2) [29,30]. The challenge we have with all these techniques is understanding the unknown bio burden of an environmental sample. Techniques are rapidly developing, looking at the Microbiome of a sample type. These are techniques which identify and semi-quantify the organisms which exists within a sample type. However, understanding their relationship to one another and how they communicate, as well as how they interact with other hosts, such as humans, still remains an area of active research (Figure 3).
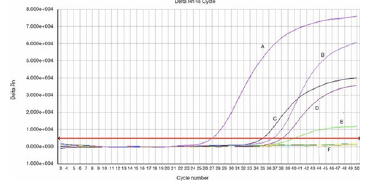
Figure 3: Real-Time PCR (RT-PCR) graph depicting the amplification curves for a number of different pathogens (A-E), which are translated into Cycle Threshold (CT) values. A positive reaction is detected by accumulation of a fluorescent signal. The CT is defined as the number of cycles required for the fluorescent signal to cross the threshold (e.g. exceeds background level).
Bacteria Specific Rapid Metabolic Assay (BSRMA)
This test allows surface, water and air testing for estimation of bacterial bio burden/ load with increased sensitivity and specificity compared to culture techniques [31-33]. Within a few minutes of sampling, the assay measures only the live bacteria present and requires no culture procedures. This type of test should not be confused with the standard total ATP (Adenosine Tri Phosphate) tests used in some food manufacturing facilities, which due to the lack of any relationship between total ATP and bacterial ATP, have no diagnostic value. There are multiple recent studies regarding the efficacy of bacteriophage-related lytic enzymes (BPLE) or viruses that infect bacteria. By degrading the cell wall of the targeted bacteria, these lytic enzymes have been shown to efficiently lyse Gram-positive and Gram-negative bacteria [34]. Bacteriophages are viruses that kill bacteria. They do not contribute to antimicrobial resistance, are safe for humans, animals, and the environment. The current focus on BPLE’s has been on their use as anti-infectives in humans and more recently in agricultural research models. The initial translational application of lytic enzymes, however, was not associated with treating or preventing a specific disease but rather as an extraction method to be incorporated in a rapid bacterial detection assay [35].
We can trace the translational history of BPLE’s from their initial discovery in 1986 for the rapid detection of group A streptococcus in clinical specimens to evolving applications in the detection and prevention of disease in humans and in agriculture [30-31]. The three -stage test uses BPLE technology to removal of all Somatic cells (all cells except bacteria) from the sample (stage 1), leaving only living bacteria in the sample to be lysed for measuring (stage2). Luminescence is then added to the sample using the positive charge on the ATP within the cell to produce light. A direct correlation between cell numbers and amount of light produced shows a statistically proven 1 – 1 Relative Light Unit (RLU) to Colony Forming Unit count [31-32]. The BSRMA was compared with the gold standard culture techniques as part of a standard of care orthopaedic pathway within an NHS hospital. The difference in sensitivity of the two tests using the same samples was stark. It is clear from this, and other studies [36-37], that the BSRMA test method is far more sensitive than standard culture techniques and, increases the users’ ability to see the numbers of live bacteria, in areas previously determined to be bacteria free. Evidence of this is seen in a recent study [10], using the BSRMA test, at multiple time points over a 4-day period.
One of the most interesting findings in the study was the levels of contamination seen immediately after the finish of the operating list, after standard night-time cleaning with Sodium Hypochlorite, and then again immediately before the first patient entered the operating theatre the next morning. Importantly, this study showed that bacterial bioburden on surfaces had returned to approximately the same levels as at the end of the previous days operating lists. Had just culture been used, levels of bacteria on surfaces at all three time points would have remained unseen.
Summary
In environmental cleaning and contamination there are two main factors associated with HAI’s, namely, the types of microorganism’s present (identification and characterisation) and the amount present (Bio burden), primarily on clinically relevant surfaces. We still do not fully understand the true interactions between, air, surfaces and hands in our healthcare facilities, but we do know they are interrelated in respect to hosting CFUs and sponsoring HAIs. Prioritising these will depend greatly on the risks attached to the hospital department, and what procedures/ work is done in those areas. Operating rooms are one of the main high-risk areas within a hospital environment for maintaining perceived aseptic conditions. There has been for many years a focus on the quality of air filtering with recommendations on issues such as air pressure, air exchanges, temperature, humidity, and filtration levels. These have been reviewed every 10-15 years since the first attempt in HTM 2025, followed by HTM 03-01 [38-39]. In the same time period, environmental cleaning and testing has for the most part been left to individual departments to decide upon, with little standardised guidance [40-43].
With the ever-increasing numbers of antibiotic resistant and disinfectant resistant bacterial species, this situation will need to change if we are to provide a safe environment for surgery to take place. Professor Dame Sally Davies (current UK Chief Medical Officer) was correct in her assessment, “if we do not do something to change the current situation of an ever-increasing risk of resistant infection post-surgery, at some point in our future we will have to stop doing what we now consider routine surgical operations as the risk from infection will be greater than the risk of not having surgery” [44]. Until we can convince organisations such as the CDC and the WHO that there is sufficient evidence to support changes, we are left with many outdated guidance documents that still impact our behaviours [45,46]. Sometimes miss quoted as being spoken first by Albert Einstein, in 1981 US Narcotics anonymously published a brochure with the phrase “Insanity is repeating the same mistakes and expecting different results”. It is clear from these small samples of data, that a lot more work is needed to accurately determine the infection risks associated with surface bacterial bioburden. Choice of the most appropriate test methodology to determine levels of bio burden is key to successful intervention.
It is certain that we must learn more from our environment, and effect change in order to improve. We should perhaps consider thinking in the same terms as Peter Drucker a management consultant who is quoted as saying, “If you can’t measure it, you can’t improve it”. Regular, fast, accurate, reliable measurement of surfaces, air and hands, has to be the goal of all healthcare facilities if we are to improve the quality of the environment in which we treat our most vulnerable patients. If we cannot achieve this goal in a timely fashion, Dame Sally Davies’s prediction of “at some point in the future”, may end up be a lot closer than anyone would like to admit. The NHS environmental SIG has drafted an Environmental Cleaning Guidance and Standards Document (ECGSD) [7] which is hoped will be the basis of a new Healthcare Technical Memoranda (HTM) looking specifically at the issues of environmental cleaning and disinfection as suggested by Dancer in 2004. The risk averse healthcare systems are now paralysed by dogma and lack of true accountability [47]. The ECGSD guide will help users to decide, where to test, which test to use, in which areas, frequency of testing with a pass/ fail test standard relevant to risk. The final guidance from this is expected to be available to trusts in the near future with standards available soon after, this is of course dependant on approval by government.
Next Steps
A microbiological environmental cleaning hospital map that colour codes risk giving guidance on all these questions to hospital infection control, microbiologists and hospital cleaners may be appropriate to help simplify the bewilderingly complex risk/ materials/ test matrix.
Final Comments
The current methods of environmental hygiene audit in hospitals and other healthcare facilities need to be supported by microbiological sampling, and further developed in line with the sample pathway previously described. The focus on process has been well described, as is the focus on the people who support it. The potential for improved changes in the behaviour of staff who know they are being observed is well understood. Goodheart’s law, sometimes confused with Heisenberg’s uncertainty principle, but which is in fact a social analogue of Heisenberg’s uncertainty principle, states “Measuring the system usually disturbs it”. In reality, the Hawthorne effect, describes a type of reactivity in which individuals modify or improve an aspect of their behaviour in response to awareness of being observed. Each variation of the theme has a common core of understanding, “If people know they are being observed, they change their behaviour”. This idea has been described many times as the “observer expectancy effect” and has been used in industry and academic institutions over many years. It may significantly underutilise as a method of achieving improvements in healthcare staff behaviour [48-51]. It could therefore have a significantly positive impact in helping to reduce the now visualisable surface bioburden that is almost certainly endemic in our healthcare facilities.
References
- Bollmann A, Lewis K, Epstein SS (2007) Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microbiol 73: 6386-6390.
- Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, et al. (2007) Manual of clinical microbiology, 9th edn, ASM Press, Washington, DC.
- Abadias M, Usall J, Anguera M, Solsona C, Viñas I (2008) Microbiological quality of fresh, minimally processed fruit and vegetables, and sprouts from retail establishments. Int J Food Microbiol 123(1–2): 121-129.
- Hoskin K (1996) The awful idea of accountability, inscribing people into the measurement of objects. International business press, London, p. 265-282
- Dancer SJ, White LF, Lamb J, Girvan EK, Robertson C (2009) Measuring the effect of enhanced cleaning in a UK hospital: a prospective cross-over study. BMC Med 7: 28-40.
- Kemp A, Laird K, Diggle M (2017) Cleaning and Disinfection Quality. Guidance standards for establishing and assessing cleaning and disinfection in UK Hospitals and other healthcare facilities.
- Abougrain AK, Nahaisi MH, Madia NS, Saied MM, Ghenghesh KS (2010) Parasitological contamination in salad vegetables in Tripoli-Libya. Food Control 21(5): 760-762.
- ISO (1996) Microbiology of food and animal feeding stuffs Horizontal method for the detection and enumeration of Listeria monocytogenes. Part 1: Detection method. ISO 11290-1. vol 1. International Organization for Standardization, Geneva, pp: 1–16.
- Russotto V, Cortegiani A, Raineri SM, Giarratano A (2015) Bacterial contamination of inanimate surfaces and equipment in the intensive care unit. J Intensive Care 10(3): 54.
- Kemp A, Hodgkinson V, Bugg A (2017) Prevention is better than cure. Clinical Services Journal 16 (9): 61-63.
- Malik RE, Cooper RA, Griffith CJ (2003) Use of audit tools to evaluate the efficacy of cleaning systems in hospitals. Am Journal Infect Control 31(3): 181-187.
- Dancer SJ (2004) How do we assess hospital cleaning? A proposal for microbiological standards for surface hygiene in hospitals. J. Hosp. Infect. 56:10-15.
- Amodio E, Dino C (2014) Use of ATP bioluminescence for assessing the cleanliness of hospital surfaces: a review of the published literature (1990-2012). J Infect Public Health 7(2): 92-98.
- Aires G, Ferreira AM, frota OP, Rigotti MA, de barcelos L, et al. (2018) Effectiveness of Surface Cleaning and Disinfection in a Brazilian Healthcare Facility. The Open Nursing Journal 12: 36-44.
- Ansari SA, Sattar SA, Springthorpe VS, Wells GA, Tostowaryk W (1988) Rotavirus survival on human hands and transfer of infectious virus to animate and nonporous inanimate surfaces. J. Clin. Microbiol 26: 1513- 1518.
- Boone SA, Gerba CP (2007) Significance of fomites in the spread of respiratory and enteric viral disease. Appl. Environ. Microbiol 73: 1687-1696.
- Cho JC, Giovannoni SJ (2004) Cultivation and Growth Characteristics of a Diverse Group of Oligotrophic Marine Gammaproteobacteria Appl Environ Microbiol 70(1): 432-440.
- Boxman IL, Dijkman R, te Loeke NA, Haqele G, Tilburg JJ, et al (2009) Environmental swabs as a tool in norovirus outbreak investigation, including outbreaks on cruise ships. J. Food Prot 72: 111-119.
- barer MR, Harwood CR (1999) Bacterial viability and culturability 41: 93-137.
- Fournier PE, Drancourt M, Colson P, Rolain JM, La Scola B, et al. (2013) Modern clinical microbiology: new challenges and solutions. Nat Rev Microbiol 11: 574-585.
- Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M (2002) Improved Culturability of Soil Bacteria and Isolation in Pure Culture of Novel Members of the Divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiology 68(5): 2391-2396.
- Rutala WA, Cole EC (1987) Ineffectiveness of hospital disinfectants against bacteria: a collaborative study. Infect. Control 8: 501-6.
- Cole EC, Rutala WA, Samsa GP (1988) Disinfectant testing using a modified use-dilution method: collaborative study. J. Assoc. Off. Anal. Chem 71: 1187-94.
- CDC (2008) Guideline for disinfection and sterilisation in healthcare facilities.
- Bartlett JM, Stirling D (2003) “A Short History of the Polymerase Chain Reaction”. PCR Protocols. Methods in Molecular Biology, pp. 3-6.
- Mullis KB, Erlich HA, Arnheim N, Horn GT, Saiki RK, et al. (1989) “Process for amplifying, detecting, and/or-cloning nucleic acid sequences” U.S. Patent 4,683,195.
- Saiki RK. “Amplification of a Short DNA Stretch by Repeated Cycles of In Vitro DNA Polymerization” American Association for the Advancement of Science.
- Erlich HA (1989) “Polymerase Chain Reaction”. Journal of clinical immunology 9 (6): 437-47.
- Handelsman J (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68(4): 669-685.
- Benschop K, Molenkamp R, van der Ham A, Wolthers K, Beld M (2008) Rapid detection of human parechoviruses in clinical samples by real- time PCR. J Clin Virol 41: 69-74.
- GR Siragusa, CN Cutter (1995) Microbial ATP bioluminescence as a means to detect contamination on artificially contaminated beef carcass tissue. Journal of food protection 58 (7): 764-769.
- CN Cutter, WJ Dorsa, GR Siagusa (1996) A rapid microbial bioluminescence assay for meat carcasses. Dairy, food and environmental sanitation 16 (11): 726-736.
- Kemp A (2018) A New era in Bacterial detection for healthcare? Clinical Services Journal 17(9): 38-39.
- Stephen T Abedon (2014) Phage therapy: Past, Present and Future. Frontiers in Microbiology 35.
- Mc Grath S, Van Sindern D (2007) Bacteriophage. Genetics and microbiology.
- Kemp A, Hodgkinson V, Bugg A (2018) Alcohol gels, more harm than good? Clinical Services Journal Vol 17(2): 44-46.
- Kemp A, Hodgkinson V, Bugg A (2018) Alcohol gels, end of an era? Clinical Services Journal 17(6) :52-54.
- Healthcare Technical Memorandum 03-01 (2007) Specialised ventilation in Healthcare premises.
- Healthcare Technical Memorandum 2025 (1994) Ventilation in Healthcare premises.
- CDC (1999) guidance on prevention of surgical site infections.
- AFPP publications - Standards and recommendations for safe Perioperative practice
- WHO (2009) guidelines for safe surgery?
- Health Protection Scotland (2016) Healthcare Associated Infection Annual report.
- Davies S. Antimicrobial resistance poses ‘catastrophic threat’, says Chief Medical Officer.UK
- WHO guidance on hand hygiene in healthcare.
- The National Specifications for cleanliness in the NHS (2007) a framework for setting and measuring performance outcomes.
- Hoskin K (1996) The awful idea of accountability, inscribing people into the measurement of objects. London, International business press 265- 282.
- Goodheart C. Goodheart’s law. Pears Cyclopedia 99th Edition 1990-1 pp: G27-31.
- Landsberger HA. The Hawthorne effects.
- Strathern M (1997) “Improving ratings” audit in the British university system. European review 5: 305-321.
- Heisenberg W. Uncertainty principle in mathematical inequalities.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.