Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Representing and Connecting the Biomedical Science through the Applied Ontology Discipline
*Corresponding author: Mauricio B Almeida, Federal University of Minas Gerais, Belo Horizonte, Brazil.
Received: February 05, 2020; Published: February 26, 2020
DOI: 10.34297/AJBSR.2020.07.001182
Abstract
Several obstacles remain for Biomedicine in reusing, disseminating and sharing specialized knowledge using information systems. The Biomedical literature is inherently ambiguous, systems developers suffer from some idiosyncrasy and solipsism, and there is not an unique standard, to mention just a few problems. The lack of integration among information systems, the so-called problem of interoperability, prevents Biomedical scientists to meet the best levels of collaboration to promote the field progress. Ontologies have been adopted in the biomedical field as tool to address the massive volume of data produced each day since the last 30 or 20 years. In this paper, we emphasize the pursuit for solutions not only through technological resources, since we believe that it should include well-stablished principles for modeling provided by the discipline of Applied Ontology.
Keywords: Ontology; Information systems interoperability; Data integration; Data architecture; Biomedical applications; Biomedical terminologies
Introduction
The term “Biomedical” dates back the 1920s and the term “Biomedicine” arose ten years later. It took just a few more years for Biomedicine came to be identified as a modern science, using the expertise of the laboratory instead of being associated with the clinical experience as in traditional medicine [1]. The first initiatives towards standardization of Biomedical terminologies emerged in the 1930s, designed to enable a clear understanding of specialized terms on a worldwide scale [2]. Terminologies are employed for knowledge representation in information systems with the aim of associating terms of a scientific field, unifying and standardizing the vocabulary for purpose of communication between experts. Biomedical terminologies are a result of the need to manage the burgeoning volume of information that have been produced within Biomedicine.
In the 1980s, one can find the origin of large efforts to gather biomedical terminologies within a unified framework. Several approaches sought this unification by means of controlled vocabularies, such as the Medical Subject Headings (MeSH), the Systematized Nomenclature of Medicine (SNOMED) and a comprehensive platform called the Unified Medical Language System (UMLS). Despite the significance of these pioneering efforts, not enough attention has been devoted to some non-technical relevant aspects, including the clarity of the terms’ meaning and classification rules. Furthermore, rather than be fruitful, this growing number of Biomedical terminologies has caused many problems, since there is not an unique standard and terminologies have included idiosyncrasy and solipsism borrowed from their developers. In addition, only the use of different standards is enough to affect the quality of data in different ways [3].
This paper aims to indicate some issues and remind of alternatives that have been applied with success for representing, organizing, and retrieving knowledge in Biomedicine, namely, ontologies. We disambiguate the term “ontology” and suggest an approach based on the discipline of Applied Ontology.
The terminological issues within the Biomedical field
One can realize that even today, with the advances of information technology, several obstacles still remain relates to reuse, disseminate and share truth knowledge. An indisputable issue impacting Biomedical information systems is the inherent ambiguity of medical literature used as source, which have been already identified since the last 30 years [4, 5]. One of the first reasons for this situation is the creation of definitions for medical terms mainly based in earlier medical dictionaries resulting in informal, circular and inconsistent definitions [6, 7]. This sort of definition can be, at most, suitable for people, but not for modern automated reasoning information systems used within the scope of the Semantic Web.
Indeed, the size, complexity, and importance of the Biomedical domain require consistent representation of specialized terms that allow automated reasoning and systems integration in a level usually called interoperability. An alternative widely accepted for knowledge representation in Biomedicine is the use of principles based on philosophical foundations. Specialized terms in information systems vocabulary can be logically defined free of ambiguities and overlaps, within a structure called “ontology” [8, 9].
How ontologies can address the Biomedical terminological issues?
Ontologies have been adopted in the Biomedical field as alternative to address the massive volume of data produced each day since the last years of 1990s [10, 11]. The meaning of term “ontology” is overloaded, but ontology has at least two main meanings: in philosophy, it is a metaphysical discipline that approaches what exists and how things are related one each other; in Computer and Information Science, ontology means a software engineering artifact [12] which includes translating natural language into axioms within a knowledge representation system. In the scope of the latter, ontology supports machine-readable representations of the context aimed at automatic reasoning, in other words, ontology is the core knowledge repository for Artificial Intelligence. The discipline called “Applied Ontology” has encompassed both aforementioned meanings in introducing philosophical principles in the activity of creating rigorous information systems models.
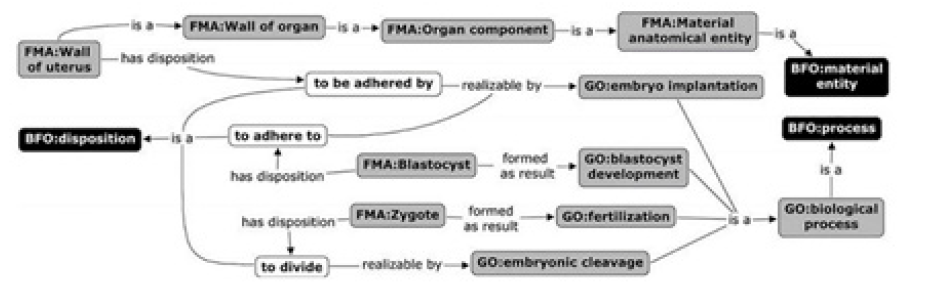
Despite we are not able to demonstrate all power of ontologies in representing Biomedical terminologies here, (Figure 1) presents a recent ontological scheme for gynecology and obstetrics specialties, still in an informal format, which outline embryo and pre-embryo stages. This scheme is part of OntoNeo [13] before the knowledge formalization required by ontology methodologies. Even in a earlier phase, it exhibits interesting properties of ontologies. As one can see, the terms inside rectangles are accompanied by labels, for example, BFO, FMA, GO. These labels stand for, respectively, Basic Formal Ontology [14], Foundation Model of Anatomy [15] and Gene Ontology [16]. While the first one is a top-level generic ontology for purposes of modeling, the second and the third are biomedical ontologies already stable, under scrutiny by scientific community for more than one decade. Thus, one can expect they have accumulated some consensual expertise within their field, in addition to follow methodologies of Applied Ontology that guarantees their suitability for XXI century information systems.
Final remarks
Issues regarding lack of interoperability among Biomedical information systems are still common, they are hampering the collaboration and eventually, the advance of Biomedical science. Ontologies take into consideration a complete framework for addressing terminological issues since they gather together, on the one hand, the most recent technologies provided by web-based inference engines; and, on the other hand, a strong set of theoretical principles, provided by philosophy, to reduce the effects of human idiosyncrasy in information systems.
References
- Scheffler RW, Strasser BJ (2015) History and Sociology of Biomedical Sciences. In: International Encyclopedia of the Social & Behavioral Sciences 2nd
- Smith B (2008) New desiderata for biomedical ontologies. In: K Munn, B Smith (Eds.), Applied ontology: an introduction. Berlin: Verlag, Germany.
- Somayeh Sadat Seyedi, Azamossadat Hosseini, Reza Rabiei, Farkhondeh Asadi, Hamid Moghaddasi (2019) Infertility Information System with an Approach to Data Architecture: A Systematic Review. Am J Biomed Sci & Res 5(4): 254-261.
- Aspevall O, Karlsson D, Forsum U (2001) Building a concept system to structure the contents of a decision support system--a grounded theory study of concepts in the knowledge domain of urinary tract infection. Med Inform Internet Med 26(2): 115-129.
- Liss P, Aspevall O, Karlsson D, Forsum U (2003) Terms used to describe urinary tract infections – the importance of conceptual clarification. APMIS 111(2): 291-299.
- Munn K, Smith B (2008) Applied ontology: An introduction. Berlin: Verlag, Germany.
- Smith B (2013) Introduction to the Logic of Definitions.
- Guarino N (1998) Formal ontology in information systems.
- Smith B (2003) Ontology. In: L M Floridi (Ed) The Blackwell guide to the philosophy of computing and information. Oxford: Wiley Blackwell.
- Rector A, Rogers J (2005) Ontological and practical issues in using a description logic to represent medical concept systems: Experience from GALEN. School of Computer Science the University of Manchester Preprint Series CSPP-35.
- Rosse C, Mejino JLV (2003) A reference ontology for biomedical informatics: The foundational model of anatomy. Biom Inform (36): 478-500.
- Gruber T (2007) Ontology of Folksonomy: A Mash-up of Apples and Oranges. International journal on Semantic Web and information systems 3(1): 1-11.
- Farinelli F, Almeida MB, ElkinPL, Smith B (2016) Dealing with Social and Legal Entities in the Obstetric and Neonatal Domain. Biological Ontology and BioCreative, CEUR Workshop proceedings 1747.
- Smith B (2015) BFO 2.0 specification and user’s guide.
- Agoncillo AV, Mejino JL V, Rickard K L, Detwiler LT, Cornelius R (2003) Proposed Classification of Cells in the Foundational Model of Anatomy. In: American Medical Informatics Association Fall Symposium. 775.
- The Gene Ontology Consortium (2008) The Gene Ontology project in 2008. Nucleic Acids Research 36 (Database issue): D440-444.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.