Research article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Epigenetic Role of Noncoding RNAs in the Recurrence of Pituitary Adenoma after Surgical Resection
*Corresponding author: Young Zoon Kim, Division of Neuro Oncology and Department of Neurosurgery, Samsung Changwon Hospital, Sungkyunkwan University School of Medicine, 158 Paryong-ro, Masanhoewon-gu, Changwon 51353, Korea.
Received: September 14, 2020; Published: October 08, 2020
DOI: 10.35097/AJBSR.2020.10.001533
Abstract
Background: Due to the lack of related symptoms and the hypersecretion of serum hormones, it is difficult to monitor and predict the postoperative recurrence of non-functioning pituitary adenomas (NFPAs). Long noncoding RNAs (lncRNAs) and protein-coding genes (PCGs) play critical roles in the development and progression of many tumors. However, the complex network of RNA interactions related to the mechanism and function of the postoperative recurrence of NFPA is still unclear.
Methods: In the present study, 100 patients with NFPA were investigated by high-throughput sequencing and follow up. Among them, 16 NFPA patients experienced recurrence at different intervals of more than five years or less than one year.
Results: By performing differential expression analysis of the fast recurrence and slow recurrence groups (t test P < 0.05), we obtained a set of differentially expressed PCGs and lncRNAs. We then identified protein-protein interaction (PPI) coregulatory networks and lncRNA-mRNA coexpression networks. In addition, we further screened the hub lncRNA-mRNA modules related to NFPA recurrence. These modules identified transcriptome expression markers for NFPA regression (log-rank test P <0.05). Finally, we evaluated the ability of the hub and module genes to predict recurrence and progression-free survival (PFS) in NFPA patients. To confirm the credibility of the bioinformatic analysis, NOL6 was randomly selected from the prognosis genes for validation by quantitative real-time polymerase chain reaction (qRT-PCR) in another set of NFPA samples (n=9).
Conclusion: These results may be helpful for evaluating the slow and rapid recurrence of NFPA after surgery, may help us explore the mechanisms of NFPA recurrence and may also serve as future effective biomarkers and therapeutic targets.
Keywords: Non-functioning pituitary adenoma, recurrence, protein-coding genes, Long noncoding RNAs
Abbreviations: NFPA: Non-Functioning Pituitary Adenoma; PCGs : Protein-Coding Genes; lncRNAs: Long noncoding RNAs; PFS: Progression-Free Survival; DEGs: Differentially Expressed Genes; DEGLs: Differentially Expressed PCGs and lncRNAs; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Introduction
Pituitary adenomas are benign neuroendocrine tumour’s [1,2] that originate from adenohypophyseal cells and account for 10-20% of intracranial neoplasm [3,4]. Pituitary adenomas can be divided into functional adenoma and non-functional adenoma according to clinical and biochemical characteristics [5]. Non-functional pituitary adenomas (NFPAs) are the most common, accounting for 14-54% of pituitary adenomas [6]. NFPA is often characterized by oppressive symptoms because it does not exhibit symptoms associated with excessive hormone production [5]. Because of the mass effect on surrounding structures, it may cause headache, visual defects, and/or hypopituitarism [6,7]. Surgical resection is the primary treatment for NFPA, but patients are often faced with tumour residue because the tumour may have invaded the cavernous sinus or surrounded the internal carotid artery [8,9]. It has been reported that approximately 12-58% of NFPA patients with macroadenoma may experience regrowth within five years [10-13]. Radiotherapy is often recommended for patients with tumour residue, but its longterm complications, such as visual defects and hypopituitarism, are still of concern [14,15]. Therefore, surgery is still the best option for patients with tumour recurrence. Serum hormone monitoring is used as a detection approach in functional pituitary adenoma; however, the absence of an effective evaluation approach for NFPA results in the failure of early intervention. Research concerning the molecular mechanisms of tumour recurrence and effective prognosis prediction methods is of great significance. Increasing evidence shows that protein-coding genes (PCGs) are involved in the activation of pathways or key proteins and play vital roles in the biological processes of pituitary adenomas. Studies by Uraki S et al. [16] show that reducing the expression of MSH6 and MSH2 can directly promote the growth of pituitary tumour’s through the ATR-Chk1 pathway [16]. Ruiqing Long et al. [17] suggested that COL6A6 interacted with P4HA3 to inhibit pituitary adenoma cell growth and invasion by inhibiting the PI3K-Akt pathway [17]. It has been reported that the low expression of TGF-β RII may be related to the development and invasion of NFPAs [18]. The research of Zhu H et al. [19] confirmed that the expression of TGF-β1 and WIF1 in recurrent tumour’s is higher than that in primary tumour’s, suggesting that these PCGs may be related to cell proliferation and recurrence [19]. Compared to noninvasive NFPAs, the expression levels of WIF1 and sFRP4 were reduced in invasive NFPAs, and WIF1 may be a potential biomarker for the aggressiveness of NFPAs [20].
Long noncoding RNAs (lncRNAs) play an important role in regulating gene expression through epigenetic or posttranscriptional mechanisms. However, lncRNAs cannot encode protein; they are a type of RNA molecule with a transcript longer than 200 nucleotides [21-23]. The differential expression and dysregulation of lncRNAs is believed to be involved in carcinogenesis and cancer progression, recurrence, and metastasis [21]. However, the role of lncRNAs in NFPA recurrence and the regulation of cellular processes remain unknown. Studies have shown that LINC00858 plays a tumourpromoting role in colon cancer by upregulating HNF4α and downregulating WNK2 [24]. Xu H et al. [25] showed that the over expressions of the lncRNA PAXIP1-AS1 can upregulate KIF14, thereby enhancing human umbilical vein endothelial cell migration, invasion, and angiogenesis in gliomas [25]. Moreover, many studies found that identifying novel lncRNA-mRNA networks by microarray analyses could contribute to exploring the potential molecular mechanisms and prognosis of tumour’s [26-28]. The above studies indicated that the dysregulation of lncRNAs and lncRNA-mRNA interactions may affect the prognosis of NFPAs.
In this study, we obtained 299 differentially expressed PCGs (228 unregulated and 71 down regulated PCGs) and 214 differentially expressed lncRNAs (120 unregulated and 94 down regulated lncRNAs) by performing differential expression analysis of the fast recurrence and slow recurrence groups (P < 0.05). We also identified protein-protein interaction (PPI) networks and coregulatory networks between lncRNAs and mRNAs. We further screened the hub lncRNA-mRNA modules related to NFPA recurrence and assessed the enrichment of the differentially expressed genes (DEGs) in different pathways by gene set enrichment analysis (GSEA). In addition, we evaluated the ability of the hub and module genes (NOL6, CDK15, MOV10, SAMM50, COL24A1, EPHX1, and DCP1A) to predict recurrence and progression-free survival (PFS) in NFPA patients. These results may help us explore the mechanisms of NFPA recurrence and may also serve as future effective biomarkers and therapeutic targets.
Materials and Methods
Patients and samples
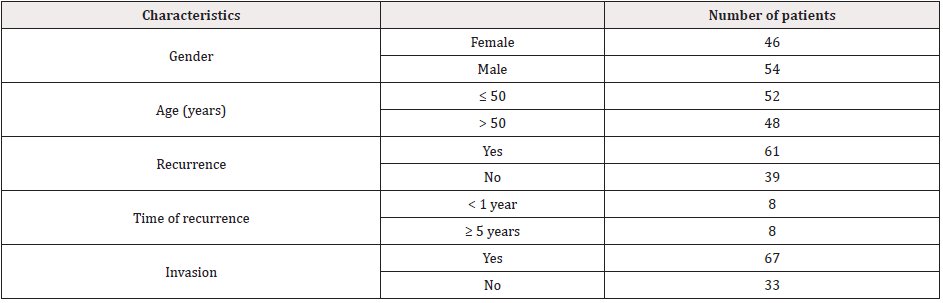
Between 2007 and 2014, 100 patients with NFPA (54 males) who underwent surgery at Samsung Changwon Hospital were recruited. The minimum follow-up time was 4 months, and the median follow-up time was 60 months (range 4-98 months). The clinicopathological characteristics of all patients are shown in Table 1. All tumour samples were immediately placed into a sample tube, frozen in liquid nitrogen and stored. Among them, 16 NFPA patients experienced recurrence at different intervals of more than five years (n=8) and less than one year (n=8). In addition, tissues from another 5 NFPAs that recurred within one year and 4 NFPAs that recurred after more than five years were collected as validated samples from patients who had undergone surgery at Samsung Changwon Hospital between 2009 and 2014. The postoperative recurrence of NFPA refers to the increase in the maximum tumour diameter measured from any direction on magnetic resonance imaging (MRI) by more than 2 mm from the day of surgery to the end of follow-up. The tumour type was determined by the patient's preoperative laboratory hormones, biochemical examination results, and postoperative pathological diagnosis.
The institutional review boards of the Samsung Changwon Hospital approved this study (SCMC 2019-05-002). All analyses were conducted according to the guidelines of the Declaration of Helsinki for biomedical research. The requirement of obtaining informed consent was waived owing to the retrospective nature of the study and minimal hazard to the participants.
Total RNA Extraction and RNA Microarray
The Phenol-free mirVana™ miRNA Isolation Kit (Cat # AM1561, Ambion, Austin, TX, USA) was used to extract and purify total RNA to generate fluorescently labeled cRNA targets (4 × 180 K). The labeled cRNA targets were then hybridized to a glass slide. After hybridization, the slides were scanned on an Agilent microarray scanner (Agilent Technologies, Santa Clara, California, USA). After extracting data using Feature Extraction software 10.7 (Santa Clara, California, USA), the Quantum algorithm was used to normalize the raw data using the Limma software package of the R program.
Identification of Differentially Expressed lncRNAs and mRNAs
Differential gene expression analysis was performed within one year after the initial postoperative NFPA (n=6) and five years later (n=6), and significance analysis of microarrays (SAM) was performed to identify the differentially expressed PCGs and lncRNAs (DEGLs) between the two groups [29]. We first downloaded the Biobase, multtest and siggenes packages from Bioconductor (http://www.bioconductor.org/). Subsequently, the available data were analyzed by the R program (www.r-project.org), and DEGLs with fold changes of > 2 and <-2 and P values of <0.05 were selected for further research.
Construction of a PPI Network and lncRNA-mRNA Coexpression Network
Cytoscape software was used to construct, visualize, and analyze the PPI network [30]. The latest version of the validated human PPI dataset was downloaded from both the Human Protein Reference Database (HPRD) (http://www.hprd.org/) (Release 9) and BioGRID (http://thebiogrid.org/) (Release 3.4.140) [31,32]. These two datasets contain 18,595 unique proteins and 174,552 interactions and were used as parent PPIs in this study. Their reliability has been effectively verified, and they have been used extensively in disease research involving human PPI networks. The nonredundant interactions in Homo sapiens from these two data sets were manually integrated [33].
First, a PPI subnetwork was generated by mapping all the DEGs and extracting them from the PPI network. To improve reliability, network reconstruction was limited to the first interacting protein neighbors of these DEGs. Second, the DEGs-adjacent protein axis was detected, and a DEGs-central PPI network was constructed. Third, after mapping all DEGs to the PPI network to detect internal interactions between the DEGs, Cytoscape was used to select nodes with all edges to create a subnetwork. The single-node and self-interactions of proteins in these subnets were deleted. The Pearson correlation test was used to calculate the coexpression relationships between lncRNAs and PCGs, and the coexpression relationships with a P value <0.05 and an absolute value of Pearson coefficient > 0.9 were selected. Finally, we obtained a lncRNA-mRNA network related to NFPA composed of differential genes. The PPI-lncRNA network was visualized using Cytoscape, and Molecular Complex Detection (MCODE) was used to identify important modules in the PPI network. We further analyzed and visualized the key modules and hub genes by using the MCODE plugin in Cytoscape. We defined the screening criteria for module genes as degree cutoff = 2, node score cutoff = 0.2, k score = 2, and maximum depth = 100.
Functional Enrichment Analysis
The ClueGO [34] plugin of Cytoscape (version 3.2.3) was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the DEGs to predict the biological functions of the prognostic lncRNAs. Functional annotations with P <0.05 were considered important. In addition, GSEA was used to identify the relevant pathways of the selected genes. We performed GSEA by using the GSEA software. The gene set used in this study was downloaded from the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp, MSigDB v4.0, released Jun 7, 2013). The Molecular Signatures Database contains various types of gene sets. The online pathway database includes 1,320 canonical pathways derived from pathway databases such as BioCarta, KEGG, Pathway Interaction Database (PID), and Reactome [35].
Validation and Efficacy evaluation of the Hub Genes by Survival Analysis
Among the hub genes, genes of interest that have not been studied in NFPA were further validated in two groups (recurrence in less than one year and recurrence after more than five years). The PFS analysis of the hub genes and module genes was performed using Kaplan-Meier curves in the R program. A P value < 0.05 was considered statistically significant.
Validation of Gene Expression by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
We used qRT-PCR with another set of NFPA samples to verify the credibility of the bioinformatics analysis. Total RNA of validated samples was extracted and purified as described above. Reverse transcription into complementary DNA (cDNA) was performed using a High Capacity cDNA Reverse Transcription Kit (0049472, Thermo Fisher) following the manufacturer's instructions. Then, Power SYBR™ Green PCR Master Mix (4367659, Thermo Fisher) was used for qRT-PCR with a total reaction volume of 20 μL. GAPDH was used as an internal control gene. All primers were synthesized by Sangon Biotech (Shanghai, China). The level of mRNAs was determined on Quant Studio 3 and 5 Systems (Applied Biosystems). For relative quantitation, expression levels were calculated using CT values (corrected for GAPDH expression) according to the equation: 2-△CT [△CT = CT (gene of interest) - CT (GAPDH)]. All qRT-PCR analyses were performed in triplicate. The sequences of the primers are as follows: GAPDH-F: 5’-ACCCACTCCTCCACCTTTGA-3’; GAPDH-R: 5’- CCACCCTGTTGCTGTAGCCA-3’; NOL6-F: 5’- ATTCGGGAAGCTGTGGTCTG-3’; NOL6-R: 5’- ATGTCAGCATGGAGTGCCAA-3’.
Results
Identification of Degls between the Fast Recurrence and Slow Recurrence Groups
Through microarray sequencing of 100 NFPA samples, we identified 18827 PCGs and 19740 lncRNAs with an expression value > 0. We analyzed the differences in PCGs and lncRNAs between 8 cases of NFPA recurrence within one year after surgery and 8 cases of NFPA recurrence five years after surgery by the SAM test. By selecting the threshold |fold change| > 2 or adjusted P <0.05, we identified a total of 299 differentially expressed PCGs (228 upregulated and 71 downregulated PCGs) and 214 differentially expressed lncRNAs (120 upregulated and 94 downregulated lncRNAs) (Figure 1A & 1B). The expression heat map further validated the results, and Figure 1C & 1D shows the differential PCGs and lncRNAs, showing different expression trends for recurrence.
Pathway Enrichment of the Degs by GSEA Classified the Fast Recurrence and Slow Recurrence Groups
We conducted GSEA and found that 30 different pathways related to 299 differentially expressed PCGs were downregulated or upregulated according to the recurrence rate. Some enriched terms are listed in Figure 2, such as regulation of cell death, regulation of cell adhesion, positive regulation of biosynthetic process, and positive regulation of gene expression, small molecule metabolic process, and response to extracellular stimulus. These results indicate that changes in these pathways lead to the recurrence and progression of NFPAs.
Dysregulated lncRNA-mRNA Interaction Network Establishment and Module Analysis
Based on the Pearson test, we constructed a differential expression network of lncRNAs and mRNAs, selecting genes with P < 0.05 and Pearson coefficient absolute value > 0.9/< 0.9 (lncRNA/mRNA quantity = 78/104, Figure 3A), and transferred this network to the differential PCG PPI parent network (see Method). Afterwards, we obtained the lncRNA-mRNA interaction network by combining these two networks. The lncRNA-mRNA network we constructed for the DEGLs contains a total of 4,490 nodes and 6,933 interactions. Figure 3B shows that the degrees of the nodes follow a power-law distribution, further illustrating that the network is similar to most biological networks, and the network is scale-free. We also calculated the average path length of the network, which shows that the characteristic path length of the network is much longer than the path length of the random network (1000 times longer than the random network, P < 0.001, Figure 3C), which implies that the network had reduced global efficiency.
Many studies have shown that PCGs and lncRNAs usually function by participating in functional modules. Through cluster analysis of the PPI network using the MCODE plugin of Cytoscape, we obtained 8 important modules according to the degree of importance. Module 1 contains 46 nodes and 54 edges (Figure 4A); Module 2 contains 71 nodes and 79 edges (Figure 4B); Module 3 contains 59 nodes and 65 edges (Figure 4C); Module 4 contains 59 nodes and 61 edges (Figure 4D); Module 5 contains 55 nodes and 55 edges (Figure 4E); Module 6 contains 44 nodes and 44 edges; Module 7 contains 83 nodes and 88 edges; and Module 8 contains 214 nodes and 218 edges.
ClueGO was used for enrichment analysis of the genes in these modules. As shown in Figure 4 (right), GO analysis indicated that the genes in Modules 1-5 were mainly concentrated in T cell migration, chemotaxis, activation, regulation of cell-cell adhesion, and regulation of cytokine production involved in the immune response and so on. In addition, KEGG analysis showed that enrichment of these module genes was mainly observed for the cell cycle, adherent junction, TNF signaling pathway, TGF-beta signaling pathway and so on. The expression of 21 hub genes and 10 module lncRNAs from the above 8 modules (CCR1, CCL3, CCL4, PACSIN1, CGNL1, TRIM69, STAB2, ATP8A1, CD48, NOL6, ZFP36, KLF4, CD247, REEP6, SQRDL, KCNJ6, ANXA2, SPRY2, KCNS3, ITM2C, THBS2, CTD-2515H24.2, LL21NC02-21A1.1, LOC200772, RP11-402C9.1, LINC01203, RP11-479G22.8, RP11-615I2.1, RP1-249I4.2, RP11-116N8.2, and RP11-288L9.4) were significantly different in the different recurrence time groups (Log2(fold change) ≥1, P<0.05, Figure 5). These results indicate that lncRNAs may regulate the downstream pathways in NFPA through gene modules and play an important role in tumour recurrence.
Evaluation of the Hub and Module Genes for Predicting the Recurrence and PFS of Patients
Then, the predictive ability of the module genes for the recurrence process was evaluated. Kaplan-Meier analysis of the central or module genes NOL6, CDK15, MOV10, SAMM50, COL24A1, EPHX1, and DCP1A showed that the patients could be divided into a high-risk group (n = 37) and a low-risk group (n = 36) according to the median value of gene expression as the cutoff value. Compared with low-risk patients, the PFS time of high-risk patients was significantly shorter (log-rank test P < 0.05, Figure 6A-6G).
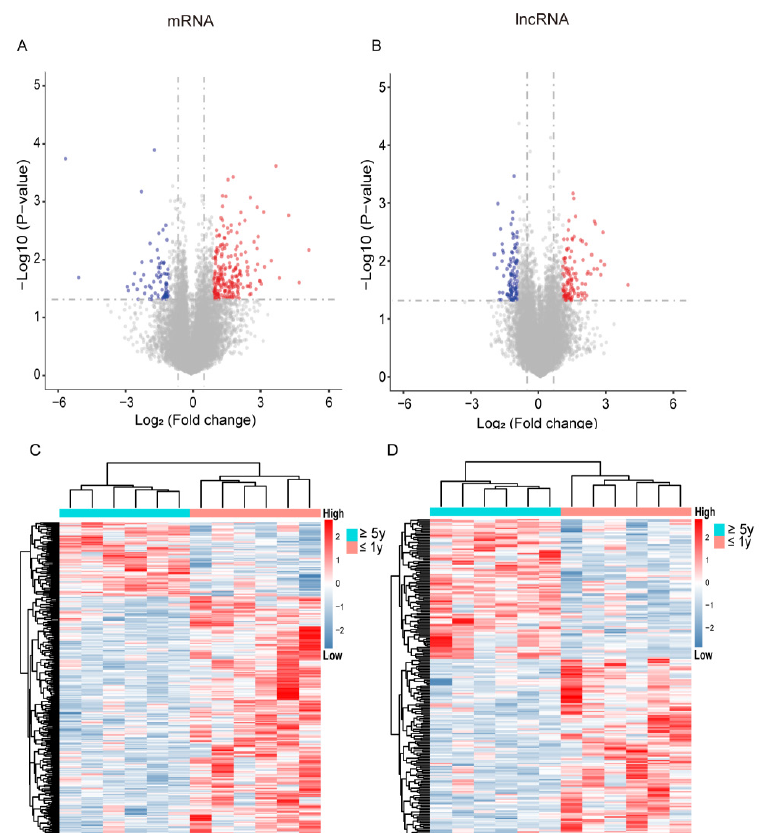
Figure 1: Identification of DEGLs related to recurrence in more than five years or less than one year. (A, B) Volcano plot of mRNAs and lncRNAs. Red slashes indicate genes without significantly different expression. Blue slashes indicate significantly regulated genes. (C, D) The expression of differential PCGs and lncRNAs in the slow recurrence group (recurrence in more than five years, ≥ 5y) or rapid recurrence group (recurrence in less than one year, ≤ 1y).
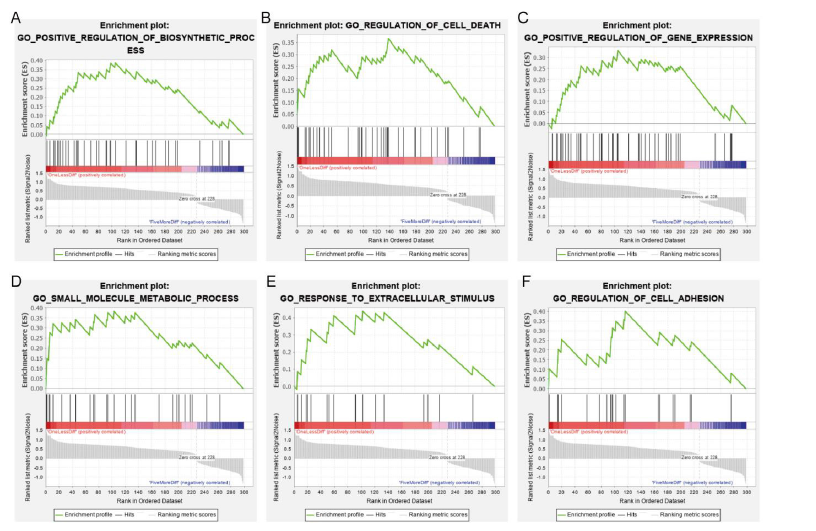
Figure 2: Pathway enrichment of the DEGs. (A-F) GSEA of DEGs between the rapid recurrence and slow recurrence groups.
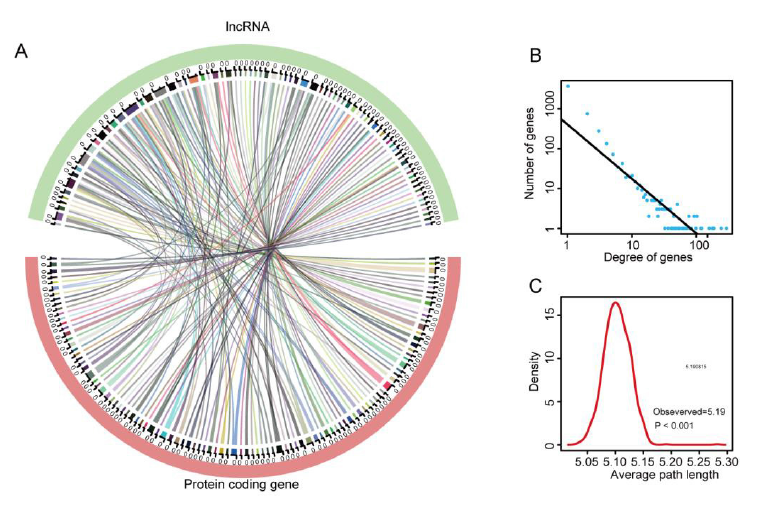
Figure 3: Topological features of the NFPA progression-related lnc RNAmRNA network. (A) The relation of dysregulated lncRNAs coexpressed with dysregulated mRNAs visualized by a Circos plot. (B) Degree distributions of the network; all degrees followed a power law distribution. (C) Average path length distributions of the real network and 1000 random networks. The average path length of the real network was larger than that of random cases.
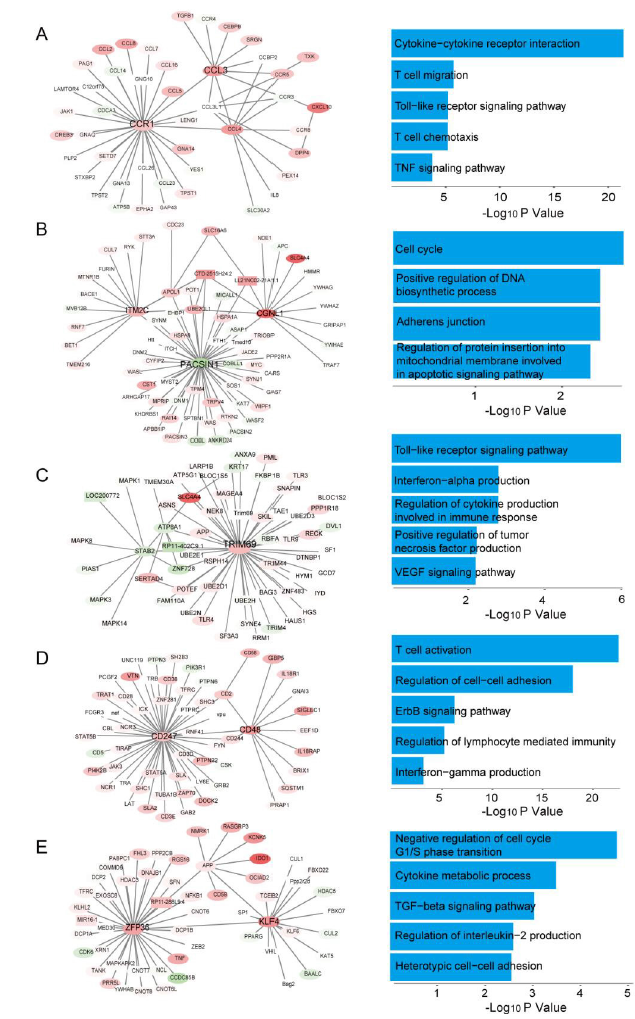
Figure 4: Module analysis of the NFPArelated lncRNAmRNA network by MCODE. The lncRNAmRNA interactions in 5 modules and pathway enrichment of PCGs in each corresponding module (AE). Square nodes represent lncRNAs, and ellipse nodes represent mRNAs (nodes in red or green: upregulation or downregulation in different NFPA progression groups).
We performed qRT-PCR to confirm the reliability of the expression profiles generated using the microarray and DEGs analysis. From among the prognostic hub and module genes above mentioned, NOL6 was randomly selected for verification (Figure 6H, P < 0.05). As expected, the qRT-PCR result basically matched the microarray analyses. These results indicate that the bioinformatics analysis of the microarray data reliably identified critical candidate genes involved in NFPA recurrence.
Discussion
NFPAs are pituitary adenomas without clinical evidence of hormonal hypersecretion. They have a prevalence of 7 to 41.34 cases/100,000, and the annual incidence is 0.65 to 2.34 cases/100,000 [6,36,37]. Transsphenoidal surgery is the recommended first-line treatment [38]. However, unlike functioning pituitary adenoma, it is difficult to monitor the tumour recurrence of NFPA through specific serum hormone alterations. When patients are re-examined because of optic nerve compression symptoms, the tumour may have grown into a large volume, which brings many obstacles to total resection and postoperative recovery. Therefore, we aimed to develop a new predictive signature that could identify early recurrence and be used as a prognostic prediction model. The main purpose of the study was to divide patients into high-risk or low-risk groups so that the most effective and timely treatment can be performed for NFPA patients.
Numerous studies have focused on the factors of tumour recurrence of NFPA to improve the prognosis of postoperative patients. Age is recognized as an important independent factor influencing the prognosis of NFPA patients, and a younger age indicates a greater chance of tumour recurrence [10,39]. However, the prognostic value of age is not as effective as the PCG and lncRNA signature in our study. Ki-67 is another commonly used pathological prognostic evaluation index [40], but a single indicator used in prognostic assessment has certain limitations in accurately evaluating the prognosis of each patient. A previous study tried to establish a statistical model that combines clinical features (age and tumour volume) and molecular markers (p16, WIF1 and TGF-β) to evaluate the recurrence probability of postoperative NFPA patients [41]. In our study, the inclusion of clinical features did not show a better efficacy. Moreover, compared with a previous study, we added a time concept to the prognostic assessment and independently assessed the prognosis of patients at different time points.
In recent years, lncRNAs have been reported in various tumour’s, serving as promising new molecular markers for tumour biological behavior, tumour diagnosis and prognostic evaluation [42,43]. The lncRNA H19 was decreased in pituitary adenomas, and its overexpression could markedly inhibit the growth of pituitary tumour cells and be used as a drug resistance marker [44]. Xing et al. [45] identified mRNAs and lncRNAs differentially expressed in clinically NFPA and normal pituitary and constructed an mRNA-lncRNA coexpression network [45]. However, their research failed to illustrate the regulatory mechanisms of the key genes or lncRNAs and their influence on patient prognosis. In this study, we focused on identifying molecular markers of NFPA recurrence.
First, we obtained the DEGLs based on NFPA recurrence in less than one year and more than five years. According to GSEA, these DEGs were enriched in the regulation of cell death and cell adhesion. Our current results are consistent with those of previous studies, which have shown that intercellular adhesion and adhesion molecules are crucial steps for tumour recurrence and proliferation [46,47]. Then, we obtained 8 modules by cluster analysis using the PPI network based on the DEGLs. GO and KEGG enrichment analyses illustrated that these module genes were mainly involved in different GO functions and pathways. For Module 1, the related GO functions were T cell migration and chemotaxis, which implies that the process of recurrence may be correlated with the immune-related tumour microenvironment. Similar to our study, Marques, P. et al. found that a low CD8:CD4 ratio is associated with a higher proliferative index (Ki-67) in pituitary adenoma [48]. In addition, KEGG analysis of other modules found that these genes were involved in the cell cycle, TNF signaling pathway, VEGF signaling pathway, TGF-beta signalling pathway and so on. These pathways might participate and regulate the proliferation and recurrence processes that occur in NFPAs.
Third, in our analysis, we obtained hub genes and module lncRNAs with significant differential expression (CCR1, CCL3, CCL4, PACSIN1, CGNL1, TRIM69, STAB2, ATP8A1, CD48, NOL6, ZFP36, KLF4, CD247, REEP6, SQRDL, KCNJ6, ANXA2, SPRY2, KCNS3, ITM2C, THBS2, CTD-2515H24.2, LL21NC02-21A1.1, LOC200772, RP11-402C9.1, LINC01203, RP11-479G22.8, RP11-615I2.1, RP1-249I4.2, RP11-116N8.2, and RP11-288L9.4). For example, Annexin A2 (ANXA2) is a pleiotropic calcium-dependent phospholipid-binding protein that is abnormally expressed in a variety of cancers [49]. Liu X et al. [50] through meta-analysis, indicated that ANXA2 overexpression might be related to poor outcomes in patients with malignant tumors [50], which is consistent with our findings. In addition, we found lncRNAs that could be used as a prognostic signature. However, the functions and regulatory mechanisms of lncRNAs in NFPA have not yet been reported.
Finally, we also assessed the predictive ability of the module genes (such as NOL6, CDK15, MOV10, SAMM50, COL24A1, EPHX1, and DCP1A) for the recurrence process. In addition, we validated the expression level of NOL6, which was randomly selected from among the hub and module genes, by qRT-PCR. The results confirmed the accuracy of our analysis.
NOL6 (nucleolar protein 6) encodes a nucleolar RNA-associated protein that is associated with the early stage of ribosome biosynthesis [51]. Dong D et al. [52] found that NOL6 is highly expressed in human prostate cancer, and knockdown of NOL6 inhibits the proliferation and mitosis and increases the cell apoptosis of human prostatic carcinoma cells (PC-3) [52]. Here, NOL6 was found to be upregulated in NFPAs recurrent within one year compared with those recurring after more than five years, suggesting that NOL6 could be a critical gene in prognostic development and a potential target for NFPA treatment. Moloney leukemia virus 10 (MOV10) belongs to the RNA helicase superfamily and could regulate mRNA stability and translation [53]. Nakano M et al. [54] found that the mRNA and protein levels of MOV10 in cancer cells were higher than those in normal cells [54]. In addition, MOV10 has been revealed to promote the angiogenesis of glioma by binding circ-DICER1 [55]. These studies indicated that MOV10 could be critical in tumorigenesis. DCP1A is a protein-coding gene for mRNA-decapping enzyme 1a, and several studies have revealed that DCP1A is upregulated in tumour tissues such as malignant melanoma, colorectal carcinoma and gastric cancer [56-58]. In addition, Tang, Y.’s study found that the high expression of DCP1A in colorectal carcinoma is correlated with poor prognosis [57], which is consistent with our results, indicating that the other PCGs and lncRNAs in our results could also be prognostic indicators for NFPA.
There are a few limitations of this study that need to be acknowledged. First, the molecular mechanisms of these PCGs and lncRNAs in NFPA are still unclear, and further studies might provide important information to understand their functional roles. Second, there are few available sequencing data about NFPA, so we were unable to verify our results in an independent validation set. Finally, the application of our signature in clinical practice should be tested prospectively. Despite these limitations of the current study, through our analysis, we verified a certain correlation between PCG and lncRNA signatures and regression. These results indicate that it is a potentially powerful prognostic marker of NFPA.
Conclusion
This is the first study to integrate PCGs and lncRNAs to predict tumor recurrence in patients with NFPA. Our study may provide a new aspect of prognostic evaluation and help patients benefit from early intervention.
Acknowledgments
The authors thank Young Min Kim, M.D. and Mi Ok Sunwoo, M.D. (Department of Radiology, Samsung Changwon Hospital) for their review of neuroradiological images, and Young Wook Kim, M.D. (Department of Biostatistics and Occupational Medicine, Samsung Changwon Hospital) for statistical advice.
Disclosure
The authors report no other conflict of interest concerning the materials or methods used in this study or the findings specified in this paper.
References
- Ostrom QT, Gittleman H, Fulop J, Max Liu, Rachel Blanda, et al. (2015) CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008-2012. Neuro Oncol 17(4): iv1-iv62.
- Fernandez A, Karavitaki N and Wass JA (2010) Prevalence of pituitary adenomas: a community-based, cross-sectional study in Banbury (Oxfordshire, UK). Clinical Endocrin 72(3): 377-382.
- Asa SL and Ezzat S. (1998) The cytogenesis and pathogenesis of pituitary adenomas. Endocrine Reviews 19(6): 798-827.
- Daly AF, Rixhon M, Adam C, Dempegioti A, Tichomirowa MA, et al. (2006) High prevalence of pituitary adenomas: a cross-sectional study in the province of Liege, Belgium. J Clin Endocrinol Metab 91(12): 4769-4775.
- Dekkers OM, Pereira AM and Romijn JA (2008) Treatment and follow-up of clinically nonfunctioning pituitary macroadenomas. J Clin Endocrinol Metab 93(10): 3717-3726.
- Ntali G and Wass JA (2018) Epidemiology, clinical presentation and diagnosis of non-functioning pituitary adenomas. Pituitary 21(2): 111-118.
- Chen L, White WL, Spetzler RF, Xu B (2011) A prospective study of nonfunctioning pituitary adenomas: presentation, management, and clinical outcome. J Neurooncol 102(1): 129-138.
- Shomali ME and Katznelson L (2002) Medical therapy of gonadotropin-producing and nonfunctioning pituitary adenomas. Pituitary 5(2): 89-98.
- Meij BP, Lopes MB, Ellegala DB, Alden TD, Laws ER (2002) The long-term significance of microscopic dural invasion in 354 patients with pituitary adenomas treated with transsphenoidal surgery. J Neurosurg 96(2): 195-208.
- Brochier S, Galland F, Kujas M, Gaillard S, Raftopoulos C, et al. (2010) Factors predicting relapse of nonfunctioning pituitary macroadenomas after neurosurgery: a study of 142 patients. Eur J Endocrinol 163(2): 193-200.
- Ferrante E, Ferraroni M, Castrignano T, Menicatti L, Anagni M, et al. (2006) Non-functioning pituitary adenoma database: a useful resource to improve the clinical management of pituitary tumors. Eur J Endocrinol 155(6): 823-829.
- Greenman Y, Ouaknine G, Veshchev I, Reider G, Segev Y, et al. (2003) Postoperative surveillance of clinically nonfunctioning pituitary macroadenomas: markers of tumour quiescence and regrowth. Clin Endocrinol 58(6): 763-769.
- Dekkers OM, Pereira AM, Roelfsema F, Voormolen JHC, Neelis KJ, et al. (2006) Observation alone after transsphenoidal surgery for nonfunctioning pituitary macroadenoma. J Clin Endocrinol Metab 91(5): 1796-1801.
- Brada M, Jankowska P (2008) Radiotherapy for pituitary adenomas. Endocrinol Metab Clin North Am 37(5): 263-275.
- Pollock BE, Cochran J, Natt N, Brown PD, Erickson D, et al. (2008) Gamma knife radiosurgery for patients with nonfunctioning pituitary adenomas: results from a 15-year experience. Int J Radiat Oncol Biol Phys 70(5): 1325-1329.
- Uraki S, Ariyasu H, Doi A, Kawai S, Takeshima K, et al. Reduced Expression of Mismatch Repair Genes MSH6/MSH2 Directly Promotes Pituitary Tumor Growth via the ATR-Chk1 Pathway. J Clin Endocrinol Metab 103(3): 1171-1179.
- Long R, Liu Z, Li J, Yu H (2019) COL6A6 interacted with P4HA3 to suppress the growth and metastasis of pituitary adenoma via blocking PI3K-Akt pathway. Aging 11(20): 8845-8859.
- Gu YH, Feng YG (2018) Down-regulation of TGF-beta RII expression is correlated with tumor growth and invasion in non-functioning pituitary adenomas. J Clin Neurosci 47: 264-268.
- Liu Z, Zhu Y, Xu L, Zhang J, Xie H,et al. (2018) Tumor stroma-infiltrating mast cells predict prognosis and adjuvant chemotherapeutic benefits in patients with muscle invasive bladder cancer. Oncoimmunology 7(9): e1474317.
- Song W, Qian L, Jing G, Jie F, Xiaosong S, et al. Aberrant expression of the sFRP and WIF1 genes in invasive non-functioning pituitary adenomas. Mol Cell Endocrinol 474: 168-175.
- Liz J, Esteller M (2016) lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta 1859(1): 169-176.
- Hu D, Su C, Jiang M, Shen Y, Shi A, et al. (2016) Fenofibrate inhibited pancreatic cancer cells proliferation via activation of p53 mediated by upregulation of LncRNA MEG3. Biochem Biophys Res Commun 471(2): 290-295.
- Zhang W, Shi S, Jiang J, Li X, Lu H , et al. (2017) LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed Pharmacother 91: 312-319.
- Xu T, Wu K, Zhang L, Zheng S, Wang X, et al. (2020) Long non-coding RNA LINC00858 exerts a tumor-promoting role in colon cancer via HNF4alpha and WNK2 regulation. Cell Oncol (Dordr) 43(2): 297-310.
- Xu H, Zhao G, Zhang Y, Jiang H, Wang W, et al. (2019) Long non-coding RNA PAXIP1-AS1 facilitates cell invasion and angiogenesis of glioma by recruiting transcription factor ETS1 to upregulate KIF14 expression. J Exp Clin Cancer Res 38(1): 486.
- Zhou S, Wang L, Yang Q, Liu H, Meng Q, et al. (2018) Systematical analysis of lncRNA-mRNA competing endogenous RNA network in breast cancer subtypes. Breast Cancer Res Treat 169(2): 267-275.
- Guo Q, Cheng Y, Liang T, He Y, Ren C, et al. (2015) Comprehensive analysis of lncRNA-mRNA co-expression patterns identifies immune-associated lncRNA biomarkers in ovarian cancer malignant progression. Sci Rep 5:17683.
- Zhang ZL, Zhao LJ, Chai L, Zhou SH, Wang F, et al. (2017) Seven LncRNA-mRNA based risk score predicts the survival of head and neck squamous cell carcinoma. Sci Rep 7(1): 309.
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98(9): 5116-5121.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11): 2498-2504.
- Goel R, Muthusamy B, Pandey A, Prasad TS (2011) Human protein reference database and human proteinpedia as discovery resources for molecular biotechnology. Mol Biotechnol 48(1): 87-95.
- Chatr Aryamontri A, Oughtred R, Boucher L, Boucher L, Rust J, et al. (2017) The BioGRID interaction database: 2017 update. Nucleic Acids Res 45(D1): D369-D379.
- Du ZP, Wu BL, Wu X, Lin XH, Qiu XY, et al. (2015) A systematic analysis of human lipocalin family and its expression in esophageal carcinoma. Sci Rep 5: 12010.
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, et al. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25(8): 1091-1093.
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102(43): 15545-15550.
- Tjornstrand A, Gunnarsson K, Evert M, Holmberg E, Ragnarsson O, et al. (2014) The incidence rate of pituitary adenomas in western Sweden for the period 2001-2011. Eur J Endocrinol 171(4): 519-526.
- Olsson DS, Nilsson AG, Bryngelsson IL, Trimpou P, Johannsson G, et al. (2015) Excess Mortality in Women and Young Adults With Nonfunctioning Pituitary Adenoma: A Swedish Nationwide Study. J Clin Endocrinol Metab 100(7): 2651-2658.
- Freda PU, Beckers AM, Katznelson L, Molitch ME, Montori VM, et al. Pituitary incidentaloma: an endocrine society clinical practice guideline. J Clin Endocrinol Metab 96(4): 894-904.
- Tampourlou M, Ntali G, Ahmed S, Arlt W, Ayuk J, et al. (2017) Outcome of Nonfunctioning Pituitary Adenomas That Regrow After Primary Treatment: A Study From Two Large UK Centers. J Clin Endocrinol Metab 102(6): 1889-1897.
- Hasanov R, Aydogan BI, Kiremitci S, Erden E, Gullu S. (2019) The Prognostic Roles of the Ki-67 Proliferation Index, P53 Expression, Mitotic Index, and Radiological Tumor Invasion in Pituitary Adenomas. Endocrinol Pathol 30(1): 49-55.
- Cheng S, Wu J, Li C, Li Y, Liu C, et al. (2019) Predicting the regrowth of clinically non-functioning pituitary adenoma with a statistical model. J Transl Med 17(1): 164.
- Chen X, Dai M, Zhu H, Li J, Huang Z, et al. (2017) Evaluation on the diagnostic and prognostic values of long non-coding RNA BLACAT1 in common types of human cancer. Mol Cancer 16(1): 160.
- Wu ZR, Yan L, Liu YT, Cao L, Guo YH, et al. (2018) Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat Commun 9(1): 4624.
- Zhang Y, Liu YT, Tang H, Xie WQ, Yao H, et al. (2019) Exosome-Transmitted lncRNA H19 Inhibits the Growth of Pituitary Adenoma. J Clin Endocrinol Metab 104(12): 6345-6356.
- Xing W, Qi Z, Huang C, Zhang N, Zhang W, et al. (2019) Genome-wide identification of lncRNAs and mRNAs differentially expressed in non-functioning pituitary adenoma and construction of an lncRNA-mRNA co-expression network. Biol Open 8(1): bio037127.
- Chen L, Liu D, Yi X, Qi L, Tian X, et al. (2020) The novel miR-1269b-regulated protein SVEP1 induces hepatocellular carcinoma proliferation and metastasis likely through the PI3K/Akt pathway. Cell Death Dis 11(5): 320.
- Ichikawa T, Okugawa Y, Toiyama Y, Tanaka K, Yin C, et al. (2019) Clinical significance and biological role of L1 cell adhesion molecule in gastric cancer. Br J Cancer 121(12): 1058-1068.
- Marques P, Barry S, Carlsen E, Collier D, Ronaldson A, et al. (2019) Chemokines modulate the tumour microenvironment in pituitary neuroendocrine tumours. Acta Neuropathol Commun 7(1):172.
- Ma S, Lu CC, Yang LY, Wang JJ, Wang BS, et al. (2018) ANXA2 promotes esophageal cancer progression by activating MYC-HIF1A-VEGF axis. J Exp Clin Cancer Res 37(1):183.
- Liu X, Ma D, Jing X, Wang B, Yang W, et al. (2015) Overexpression of ANXA2 predicts adverse outcomes of patients with malignant tumors: a systematic review and meta-analysis. Med Oncol 32(1): 392.
- Utama B, Kennedy D, Ru K, Mattick JS (2002) Isolation and characterization of a new nucleolar protein, Nrap, that is conserved from yeast to humans. Genes Cells 7(2): 115-132.
- Dong D, Song M, Wu X, Wang W (2020) NOL6, a new founding oncogene in human prostate cancer and targeted by miR-590-3p. Cytotechnology 72(3): 469-478.
- Wang W, Snyder N, Worth AJ, Blair IA and, Witze ES (2015) Regulation of lipid synthesis by the RNA helicase Mov10 controls Wnt5a production. Oncogenesis 4(6): e154.
- Nakano M, Kakiuchi Y, Shimada Y, Mika Ohyama, Yasuko O, et al. MOV10 as a novel telomerase-associated protein. Biochemical and biophysical research communications 388(2): 328-332, 2009.
- He Q, Zhao L, Liu X, Ohyama M, Ogiwara Y, et al. (2019) MOV10 binding circ-DICER1 regulates the angiogenesis of glioma via miR-103a-3p/miR-382-5p mediated ZIC4 expression change. J Exp Clin Cancer Res 38(1): 9.
- Tang Y, Xie C, Zhang Y, Qin Y, Zhang W (2018) Overexpression of mRNA-decapping enzyme 1a predicts disease-specific survival in malignant melanoma. Melanoma Res 28(1): 30-36.
- Wu C, Liu W, Ruan T, Zhu X, Tao K, et al. (2018) Overexpression of mRNA-decapping enzyme 1a affects survival rate in colorectal carcinoma. Oncol Lett 16(1): 1095-1100.
- Shi C, Liu T, Chi J, Luo H, Wu Z, et al. (2019) LINC00339 promotes gastric cancer progression by elevating DCP1A expression via inhibiting miR-377-3p. J Cell Physiol 234(12): 23667-23674.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.