Opinion 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Isochores: Three-Dimensional Information Layer in DNA Challenges Junk DNA Paradigm
*Corresponding author: Boris Schmidtgall, Department of molecular biology, The independent Research Initiative on Information & Origins, Germany.
Received: October 05, 2020;Published: October 16, 2020
DOI: 10.35097/AJBSR.2020.10.001541
Abstract
Over the past forty years, many different types of DNA structures in the genome of living organisms have been classified as “junk DNA”, i.e., non-functional genetic units that are thought to be traces of a long and protracted evolution. Gradually, however, it is becoming apparent that these genetic regions perform important functions (pseudogenes, introns, etc.). A similar case that recently has become known are DNA sequences with a frequently repeating structural known as the isochores. The finding that isochores are genetic elements that store the information for the correct spatial arrangement of DNA in the cell nucleus challenges the evolutionary junk DNA paradigm.
Opinion
Without doubt, the genetic material of all living beings (DNA) is one of the most fascinating biomolecules. Not long after characterizing its three-dimensional helix structure (1953), James Watson said: "I believe few discoveries have been of such perfect beauty." As it seems, the moments of surprise associated with the study of DNA have not been exhausted even today-after almost 70 years of intensive research. After the genetic code was deciphered and the packaging of DNA was characterized in the 1960s and 1970s, some influential experts came to the conclusion that only protein-coding DNA sequences comprising approximately 2% of the whole genome were functional. Based on the theory of evolution, the remaining 98% were dubbed "junk DNA", i.e. genetic waste that had become inoperable in the course of many millions of years of evolutionary change [1].
However, this notion has increasingly been questioned over the past 20 years, as a large number of findings suggest that at least 80% of the human genome is functional (ENCODE Consortium 2012). The main fallacy of the junk DNA hypothesis was the hasty assumption that genetic sections that are not used for the synthesis of proteins do not serve any function at all. Indeed, it has repeatedly been found that gene sequences previously thought to be functionless perform important regulatory functions. The term "pseudogene", which is often used in this context, has therefore often proven to be incorrect [2]. Similarly, introns, which were previously thought to be evolutionary remnants, have also been shown to be functional upon closer examination [3].
Another type of genetic regions, which were also classified as "junk DNA" for a long time, are the isochores. These are long, repetitive DNA segments containing a frequently recurring structural motif, which have been considered to be relics of viruses [4]. However, recently the Italian molecular biologist Giorgio Bernardi reported in a review article that the isochores apparently perform very important functions for the entire cell [5]. Bernardi found "…that these structures, essentially based on the distribution and topology of short sequences, mold primary chromatin domains (and define nucleosome binding)."
Put simply, this means that the isochores, depending on their structural makeup, significantly influence the 3D-structure of the packaging of the DNA in the cell nucleus. Bernardi speaks in this context of a "genomic code"-i.e. coded information about how the DNA in the cell nucleus should be arranged three-dimensionally: "Furthermore, being pervasive in the genome, the genomic code solves old outstanding problems, such as 'non-coding DNA', 'junk DNA' or 'selfish DNA'". Accordingly, the effect of the isochores is by no means locally limited, but affects the entire structure of the hereditary molecules in the cell nucleus.
The isochores do not have uniform compositions and are therefore divided into five subgroups (L1, L2, H1, H2, H3). The classification into the subgroups of the isochores depends on the content of GC base pairs. GC-poor isochores are assigned to the two subgroups L1 and L2.
These genetic regions are poor in expressed sections ("gene deserts") and are mainly located on the inside of the cell wall of the cell nucleus. They represent a kind of anchor of chromatin on the wall of the cell nucleus [6]. This anchor is called Lamina-Associated Domain (LAD) and promotes the correct spatial arrangement of the chromatin in the cell nucleus (structure of LAD (Figure 1)). Isochores of the classes H2 and H3 have a significantly higher GC content. They protrude into the cell nucleus, where they form the structure of the Topologically Associated Domains (TAD) through mutual interactions (cohesin) - a function that keeps the chromatin in the correct three-dimensional shape (structure of TAD (Figure 2)). In addition, the GC-rich isochores contain significantly more protein-coding sections than the GC-poor sections of classes L1 and L2. Details of the function of LAD and TAD have not yet been thoroughly investigated, but it is known that changes in these structures can cause serious diseases (cancer) [6].
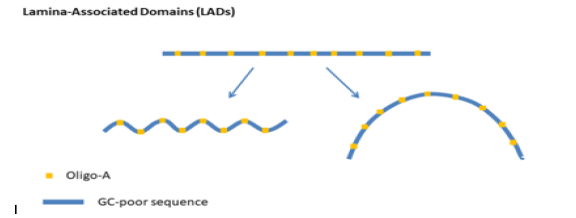
Figure 1: Schematic representation of the structural motifs of Lamina-Associated Domains (LADs). These are GC-poor genetic regions doped with oligo-A units. These units cause local curve or arc structures in the DNA.
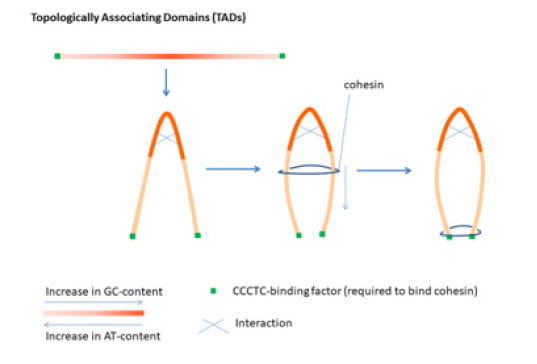
Figure 2: Model of TAD folding: Internal interactions in the GC-rich region first form a loop structure. This arrangement is then further attached by a cohesin (ring-shaped protein). The cohesin slides from the GC-rich region to the CCCTC binding factor, where it finally binds.
The fact that the same DNA sequences both encode proteins and contain genomic coding, means that there are two overlapping levels of information within these sequences. Thus, a high information density has been realized very efficiently. The presence of several overlapping information levels on the DNA has already been recognized in other contexts: Intrinsically unstructured proteins are known to adopt several specific folds-a properties that presupposes corresponding multi-layer information storage on the genetic material [7]. The extremely high information density realized on the DNA is a very reliable design indicator.
Taking a closer look to isochores can also help to explain another biochemical fact in a meaningful way: The variability of the third position of DNA codons. This can be illustrated using an example: The amino acid arginine is encoded by the nucleotide combinations CGT, CGC, CGA, CGG, i.e. the third position is redundant here. Based on the theory of evolution it was assumed that the last, variable position is unimportant for coding and a kind of hint to the undirected forces of nature, which are said to have produced the genetic code. But with the knowledge of the genomic code it becomes clear that the last position is apparently there to store the genomic code - parallel to the coding of the structure of proteins. Thus, although the same amino acid may be encoded in different genetic sections, the last position can determine, by choosing A/T or G/C, whether the genetic section is a unit that functions in the LAD or a sequence that is effective at the TAD.
Conclusion
Bernardi concludes at several points in his review article that with the discovery of the "genomic code" the evolutionary concept of "junk DNA" becomes very questionable. However, he still tries to trace the origin of genetic information back to evolution: "By the end of the 1980s, our knowledge of the isochore organization of the human genome had not only rejected what had been called the ‘bean-bag’ view of the genome, that is, a collection of genes randomly scattered over vast expanses of ‘junk DNA’; but it had also indicated that the genome is an integrated structural, functional, and evolutionary system."
The quoted sentence seems quite conclusive in itself except the word ‘evolutionary’. In this sentence, this word is completely superfluous. The term ‘evolutionary’ here does not arise in a meaningful way from what has been said before, but merely represents an ideological confession of the author. In view of the findings described, however, it is inappropriate to describe the system as ‘evolutionary’. For how can a system at the same time exhibit essential characteristics of a created object (integrated, structured, functional) and yet emerge from undirected natural processes? This dichotomy occurs particularly often in connection with discoveries in the field of molecular biology and is never resolved. This may be due to the fact that hardly any other field provides such clear indications of design as molecular biology or genetics. Therefore, in this context the evolutionary world view can only be maintained through rhetorical concealment.
It remains to be hoped that in the future scientists will be honest and courageous enough not to follow the ideological dictates of the educational and media institutions. Instead of repeated avowals to naturalism we need to focus on the scientific findings and the conclusions that necessarily follow from them.
References
- Ohno S (1972) So much "junk" DNA in our genome. In: Smith HH (ed.). Evolution of Genetic Systems. Gordon and Breach, New York, USA. pp. 366-370.
- Borger P, Schmidtgall B (2020) Free Science from Dogma. Or: How the “Pseudogene” Hampered Scientific Progress. AJBSR 8(1): 57-58.
- Parenteau J, Maignon L, Berthoumieux M, Catala M, Gagnon V, et al. (2019) Introns are mediators of cell response to starvation. Nature pp.612-617.
- Palazzo AF, Gregory TR (2014) The case for junk DNA. PLoS Genetics 10(5): 1004351.
- Bernardi G (2019) The genomic code: a pervasive coding/molding of chromatin structures. Bio Essays 41(12): 1900106.
- Lochs SJA, Kefalopoulou S, Kind J (2019) Lamina Associated Domains and Gene Regulation in Development and Cancer. Cells 8(3): 271.
- Schmidtgall B (2018) Intrinsisch unstrukturierte Proteine. Stud Integr J 25(2): 84-91.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.