Opinion 
 Creative Commons, CC-BY
Creative Commons, CC-BY
droTAG: Adapting dTAG Toolkit to Drosophila Melanogaster
*Corresponding author: Yuri Prozzillo, Department of Biology and Biotechnology “Charles Darwin”, Sapienza University of Rome, 00185 Rome, Italy
Received: January 04, 2021; Published: January 20, 2021
DOI: 10.34297/AJBSR.2021.11.001662
Opinion
Targeted protein degradation (TPD) allows an acute and reversible knockdown of protein of interest (POI) so that the direct effects of protein depletion can be studied and distinguished from secondary effects or adaptive responses [1]. Therefore, protein degradation techniques apply in studying the function of gene products in a short time frame, and rapid effectiveness can be exploited to downregulate POI in a stage-specific manner or when time is a relevant factor, including cell division.
In the last decade, several strategies have been developed to obtain an accurate and efficient protein degradation, such as deGradFP, Auxin-inducible Degradation (AID) and degradation TAG (dTAG) which aim to achieve proteolysis of POI exploiting the powerful of degradation signal peptide sequences (tags) to hijack POI to E3 ubiquitin ligases for ubiquitylation and consequentially proteasomal degradation by recruitment of the ubiquitin-proteasome system [2].
deGradFP exploits the proteasome-based pathway to achieve direct depletion of GFP-tagged proteins, while AID needs of Auxin and transgenic OsTIR1 adapter to trigger POI depletion.
Instead, the dTAG system developed by Nabet et al. [3] induces a rapid and selective degradation of POI exploiting the heterobifunctional activity of degrader (dTAG-13). This molecule binds both FKBP12F36V-fused POI and Cereblon (CRBN), the recognition unit of CRL4-CRBN E3 ubiquitin ligase complex leading to exclusive POI degradation by the proteasome.
Most of these tools are versatile and have been adapted to work in different model organisms spanning yeast (S. cerevisiae) to humans (H. sapiens). Indeed, deGradFP strategy has been firstly developed in non-vertebrates (Drosophila) by Caussinus et al. [4], and later tailored in humans [5], while the AID system evolved in the exact opposite way arising from humans and later adapted in Drosophila by Trost et al. [6].
On the other hand, dTAG system have been well established in vitro and in vivo in different species, but to date, no attempt has been made to extend it to Drosophila, as a matter of fact, Yesbolatova et al. [7] claim that CRBN is not evolutionarily conserved in non-vertebrates, therefore dTAG is likely not functional in these organisms.
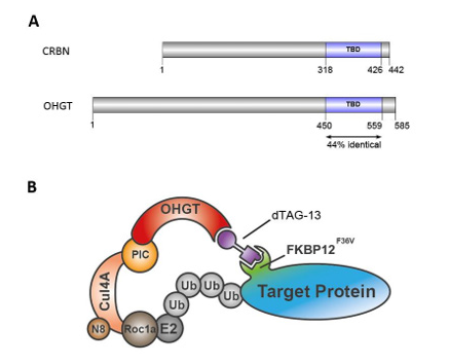
In contrast, here we would like to focus the attention on the conservation of all components of the mammalian CRL4 complex in Drosophila, including the Cullin 4 homolog and the DDB1 homolog PIC [8]. In particular, it must be emphasized that there is a 44% of aminoacidic identity in thalidomide binding domain (TBD) between human CRBN and Drosophila OHGT (Figure 1A) [9]. This data strongly supports the idea that the function of the ubiquitin ligase complex is evolutionary conserved in Drosophila melanogaster, making the hypothetical applicability of dTAG in Drosophila (droTAG), a powerful strategy that is worth to be developed (Figure 1B).
In conclusion, drawing up droTAG could be indispensable for all the worldwide fly researchers, including us, who have the possibility to highlight phenotypes that otherwise they cannot see by using conventional RNA interference approaches [10-14].
- Schemes of human Cereblon (CRBN) and its Drosophila orthologue, Ohgata (OHGT). The evolutionary conserved thalidomide binding domains (TBD) are indicated as filled boxes (blue). Percentage of identity at the amino acid level is noted.
- Cartoon showing a putative mechanism of droTAG system in Drosophila melanogaster. Heterobifunctional dTAG-13 molecules bring together OHGT and FKBP12F36V-fused POI, hijacking it towards endogenous proteasome machinery for rapid degradation. CRL4–OHGT E3 ubiquitin ligase include cullin scaffold (CUL4A), adaptor protein (PIC), substrate receptor (OHGT), N8 ubiquitin-like protein (NEDD8) and the RING protein (Roc1a) recruiting an E2 ligase.
Conflict of Interest
There is no conflict of interest
References
- Roth S, Fulcher LJ, Sapkota GP (2019) Advances in targeted degradation of endogenous proteins. Cellular and molecular life sciences 76(14): 2761-2777.
- Prozzillo Y, Fattorini G, Santopietro MV, Suglia L, Ruggiero A, et al. (2020) Targeted Protein Degradation Tools: Overview and Future Perspectives. Biology 9(12): 421.
- Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, et al. (2018) The dTAG system for immediate and target-specific protein degradation. Nature chemical biology 14(5): 431-441.
- Caussinus E, Kanca O, Affolter M (2011) Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nature structural & molecular biology 19(1): 117-121.
- Caussinus E, Kanca O, Affolter M (2013) Protein knockouts in living eukaryotes using deGradFP and green fluorescent protein fusion targets. Current protocols in protein science 73: 30.2.1-30.2.13.
- Trost M, Blattner AC, Lehner CF (2016) Regulated protein depletion by the auxin-inducible degradation system in Drosophila melanogaster. Fly 10(1): 35-46.
- Yesbolatova A, Tominari Y, Kanemaki MT (2019) Ligand-induced genetic degradation as a tool for target validation. Drug discovery today. Technologies 31: 91-98.
- Hu J, Zacharek S, He YJ, Lee H, Shumway S, et al. (2008) WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes & development 22(7): 866-871.
- Wakabayashi S, Sawamura N, Voelzmann A, Broemer M, Asahi T, et al. (2016) Ohgata, the Single Drosophila Ortholog of Human Cereblon, Regulates Insulin Signaling-dependent Organismic Growth. The Journal of biological chemistry 291(48): 25120-25132.
- Messina G, Damia E, Fanti L, Atterrato MT, Celauro E, et al. (2014) Yeti, an essential Drosophila melanogaster gene, encodes a protein required for chromatin organization. Journal of cell science 127: 2577-2588.
- Messina G, Atterrato MT, Fanti L, Giordano E, Dimitri P (2016) Expression of human Cfdp1 gene in Drosophila reveals new insights into the function of the evolutionarily conserved BCNT protein family. Scientific reports 6: 25511.
- Jonchere V, Alqadri N, Herbert J, Dodgson L, Mason D, (2017) Transcriptional responses to hyperplastic MRL signalling in Drosophila. Open biology 7(2): 160306.
- Taylor E, Alqadri N, Dodgson L, Mason D, Lyulcheva E, et al. (2017) MRL proteins cooperate with activated Ras in glia to drive distinct oncogenic outcomes. Oncogene 36(30): 4311-4322.
- Prozzillo Y, Delle Monache F, Ferreri D, Cuticone S, Dimitri P, et al. (2019) The True Story of Yeti, the "Abominable" Heterochromatic Gene of Drosophila melanogaster. Frontiers in physiology 10: 1093.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.