Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Comparison Between the SD BIOLINE Dengue Duo and Two InBios Prototype Dengue Immunochromatographic Tests
*Corresponding author: Hua-Wei Chen, Naval Medical Research Center, 503 Robert Grant Ave, Silver Spring, MD 20910, USA
Received: April 04, 2021; Published: April 19, 2021
DOI: 10.34297/AJBSR.2021.12.001772
Abstract
Dengue fever, caused by any of the four serotypes of dengue viruses (DENV1-4), is the most important arthropod-borne viral disease in the world. Although dengue diagnostic tests based on polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA) have received the U.S. Food and Drug Administration (FDA) clearance, these assays require specialized equipment and highly trained personnel, and thus not suitable for resource-limited settings. Two immunochromatographic test (ICT) prototypes (traditional and multiplex formats) were recently developed by InBios International Inc. (Seattle, WA) to detect the presence of dengue viral non-structural protein 1 (NS1) antigen or anti-DENV IgG and IgM antibodies in serum.
Here, we report a follow-up evaluation of two InBios ICT prototypes compared to the commercially available SD BIOLINE Dengue Duo rapid test (Abbott, Santa Clara, CA) with an additional panel of 130 well-characterized, de-identified clinical samples. This panel included 25 acuteconvalescent pairs of DENV3, 20 pairs of DENV4, 10 acute non-DENV febrile samples and 10 normal human sera along with 20 Zika clinical samples. Both the InBios traditional format and the SD Duo test showed the same overall sensitivity (81.1%) and specificity (80.0%). When compared to the reference testing methods. The InBios traditional format had a better overall performance (94.8% sensitivity and 87.9% specificity on DENV3 samples; 100.0% sensitivity and 80.0% specificity on DENV4 samples) than the multiplex format (93.0% sensitivity and 28.6% specificity on DENV3 samples; 86.7% sensitivity and 40.0% specificity on DENV4 samples) when compared to the SD Duo test results.
The InBios multiplex format had higher overall sensitivity (85.6%) but much lower specificity (55.5%). All three dengue rapid tests crossreacted with Zika clinical samples resulting in false positive for dengue in 20% to 73.3% of the Zika positive samples. Both the InBios traditional format and the SD Duo test had better overall performance than the InBios multiplex format. These easy and simple rapid tests with fast turn-around time are ideal for use in resource-limited settings where dengue is endemic.
Keywords: Dengue Virus, Immunochromatographic Test (ICT), Evaluation, Diagnostic, Rapid Test
Introduction
Dengue fever is the most prevalent arthropod-borne viral disease in the world and thus it is the most significant risk of arboviral disease to deployed U.S. military personnel [1]. The disease is endemic in over 100 countries around the world, including countries in Central and South Americas, the Caribbean, Southeast Asia, Africa, Eastern Mediterranean, and Western Pacific regions [2,3]. There are four serotypes present of dengue virus (DENV1-4) and infection with any of these four serotypes can result in an asymptomatic infection or a symptomatic infection whose manifestation ranges from mild self-limiting fever to severe disease with high fever, hemorrhage, and shock [4]. Dengue fever ranks third on the list of the Global Ranked List of Infectious Diseases Threats from the U.S. Military Infectious Disease Threats Prioritization Panel (ID-TPP) due to its high risk and potential impacts to military operations [5]. Efforts to mitigate associated morbidity and mortality of dengue are a high priority for the U.S. military research and acquisition [6,7].
Unfortunately, even though a licensed dengue vaccine is available for use in individuals with previous dengue infection, this option is not available for dengue naïve populations including most of U.S. military personnel. Also, there is no specific antiviral therapeutic to prevent or treat dengue infection. Hence, early and prompt diagnosis of dengue infection is crucial for monitoring patients and for alerting health care providers to provide appropriate hospitalization and supportive therapy for patients in anticipation that they may progress to manifest the severe form of the disease. There are U.S. Food and Drug Administration (FDA)- cleared diagnostic assays for dengue infection using polymerase chain reaction (PCR) and IgM enzyme-linked immunosorbent assay (ELISA) based assays [8,9]. However, these assays require specialized equipment and highly trained personnel that are not always readily available especially in resource-constrained areas such as in rural areas, developing countries, or military deployment missions.
InBios International Inc. (Seattle, WA) recently developed two immunochromatographic test (ICT) prototypes (traditional and multiplex formats) for the detection of the dengue viral nonstructural protein 1 (NS1) antigen or anti-DENV IgG and IgM antibodies in serum samples of dengue suspected patients. These rapid tests can give result within 20 minutes by naked eyes and also has the advantage of being able to be performed without any equipment.
Recently, an evaluation of two InBios ICT prototypes (traditional and multiplex formats) was performed by our group with 183 well-characterized clinical samples from a previously conducted multicenter clinical trial [10]. The traditional format showed better overall performance (sensitivity: 97.4%; specificity: 90%) compared to the multiplex format (sensitivity: 86.9%; specificity: 63.3%) (10). However, that study only used low number of DENV3 and DENV4 samples (6 and 27 samples, respectively) and no clinical samples from other flaviviruses such as Zika, which means no cross-reactivity of the rapid tests was determined. This is a follow-up evaluation of two InBios prototypes using an additional panel of 130 well-characterized DENV3, DENV4, and Zika samples.
This study aimed to further evaluate the performance of the two InBios prototypes in detecting dengue infection by testing more DENV3 and DENV4 samples along with Zika samples and to evaluate the performance of two InBios prototypes compared to the commercially available SD BIOLINE Dengue Duo (SD Duo) rapid test (Abbott, Santa Clara, CA; former Alere Inc., Waltham, MA) because of the extensive historical performance data available for the SD Duo test [11-14]. The overall aim of this study was to choose the best candidate between the two InBios ICT prototypes that will be used in a multicenter clinical trial as part of the process to achieve the FDA clearance.
Materials and Methods
Clinical samples
One hundred thirty de-identified clinical samples were chosen for this study, consisting of 25 acute-convalescent pairs of DENV3 samples, 20 pairs of DENV4 samples, 10 acute non-DENV febrile samples and 10 normal human sera (for a total of 110 samples) from the Naval Medical Research Unit Two (NAMRU-2) (Phnom Penh, Cambodia) with 20 Zika clinical samples (5 PCR positive and 15 antibody positive) from the Naval Infectious Diseases Diagnostic Laboratory (NIDDL) at the Naval Medical Research Center (NMRC) (Silver Spring, MD) for determining any cross-reactivity. After submission of the protocol entitled “Evaluation of Diagnostic Assays for Dengue Virus Infection Using Previously Collected Human Serum Samples” to the NMRC Institutional Review Board for review, the protocol was determined to not meet the definition of human subject research and thus was approved with an assigned project number (PJT-17-6).
All clinical samples were tested by an established traditional pathogen diagnostic algorithm at NAMRU-2 [15]. Basically, all of the clinical samples were first tested for influenza virus by a universal RT-PCR. If they were negative for influenza, they were then tested for malaria by the standard microscopy method, rickettsia by inhouse ELISA, dengue by PanBio ELISA kits (Abbott, Santa Clara, CA; former Alere Inc., Waltham, MA) (acute samples by IgM ELISA and convalescent samples by IgG ELISA), and pan-flaviviruses by using a universal primer set within a RT-PCR. If the sample was positive by PCR for pan-flaviviruses, it was then tested for dengue using serotype-specific PCR assays. For Zika clinical samples from NIDDL, the Trioplex real-time RT-PCR assay that was developed by the Centers for Disease Control and Prevention (CDC) (for the simultaneous detection of dengue, chikungunya, and Zika viruses) was used for Zika virus detection and IgM antibody capture ELISA followed by a Zika confirmatory plaque reduction neutralization test (PRNT) was used for the detection of antibodies to Zika [16].
Test Performance
The manufacturers’ instructions were followed in performing all the rapid tests. The InBios DENV Detect™ Multiplex Rapid Test (i.e., multiplex format) is a lateral flow immunoassay for the simultaneous and qualitative detection of the dengue NS1 antigen, IgM, and IgG antibodies against the envelope (E) protein of dengue virus. The multiplex format has one sample port and one buffer port on each device (Figure 1a). The InBios DENV Detect™ Rapid Test (i.e., traditional format) also provides rapid detection of dengue NS1 antigen and IgM and IgG antibodies separately. In contrast, the traditional format has two separate sample and buffer ports on each device (Figure 1b).
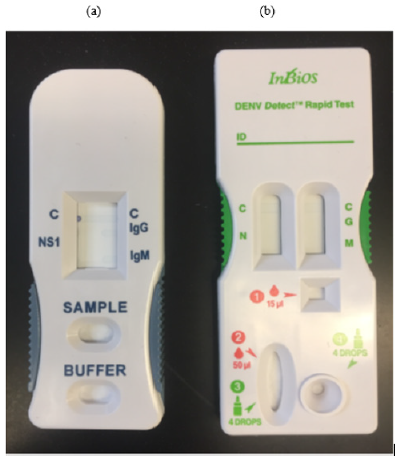
Figure 1: InBios ICT prototypes (a) Multiplex format, added 50 μl of serum to the sample loading port and followed by five drops of chase buffer to the buffer loading port, C is the control line. (b) Traditional format added 15 μl and 50 μl of serum to the sample loading ports for IgM/IgG (red drop on the right) and NS1 (red drop on the left) detection, respectively. Added chase buffer to the buffer loading port for IgM/IgG detection (green bottle on the right) and to the sample port for NS1 detection (green bottle on the left). C is the control line; N is NS1 test line; G is IgG test line; M is IgM test line. Tests were run for 20 minutes at room temperature before reading the results.
Briefly, for the InBios prototypes, 50 μl (for the multiplex format or the traditional format NS1 detection) or 15 μl of serum (for the traditional format IgM/IgG detection) was added to the sample loading port. Afterwards, five drops of chase buffer were added to the buffer loading port for the multiplex format, or the traditional format IgM/IgG detection and three drops of chase buffer were added to the buffer loading port for the traditional format NS1 detection. The results were read within a 20–25-minute reading period. For the SD Dengue Duo rapid test, 10 μl (for IgM/ IgG detection) or 100 μl of serum (for NS1 detection) was added to the sample well. Following that, four drops of assay diluent were added to the assay diluent well for IgM/IgG detection. Tests results were read within the 15-20 minutes reading period.
Data Analysis
All 130 clinical samples were tested with three rapid tests (SD Duo rapid test and both the multiplex and traditional formats of the InBios prototypes) and the results for each analyte (NS1, IgM, and IgG) for each rapid test were recorded. The sensitivity and specificity were determined as reported previously [11]. The overall or composite sensitivity was calculated for those samples that were considered positive if any of the three analytes for that particular sample was positive. Binomial method was used to calculate the confidence intervals (CIs) for sensitivity and specificity.
Results
When compared to the standard clinical diagnostic results determined by traditional diagnostic assays (including PCR and IgM/IgG ELISA) performed at NAMRU-2 as the reference standard for those 110 dengue and non-dengue samples, both the InBios traditional format and SD Duo test showed the same overall sensitivity (81.1%) and overall specificity (80.0%). The InBios multiplex format had higher overall sensitivity (85.6%) but much lower overall specificity (55.5%) (Table 1). When both of the InBios prototypes were evaluated using the SD Duo test as the reference assay, the InBios traditional format had a better overall performance (94.8% sensitivity and 87.9% specificity on DENV3 samples; 100.0% sensitivity and 80.0% specificity on DENV4 samples) than the multiplex format (93.0% sensitivity and 28.6% specificity on DENV3 samples; 86.7% sensitivity and 40.0% specificity on DENV4 samples) (Table 2a & 2b).

Table 1: Device performance using 110 serum samples from NAMRU-2 as compared to standard clinical diagnosisa.

Table 2: Device performance for InBios prototypes using (a) DENV3 (DV3) samples and (b) DENV4 (DV4) samples for composite results or for individual analyte comparing to the SD Dengue Duo results.
Both InBios formats had identical sensitivity (92.9%) and specificity (94.4%) for NS1 detection on DENV3 samples (Table 2a) and had very high sensitivity (100.0%) and specificity (multiplex format 100.0%; traditional format 96.6%) for NS1 detection on DENV4 samples (Table 2b). The traditional format had a better performance on IgG detection (81.8% sensitivity and 100.0% specificity on DENV3; 91.3% sensitivity and 88.2% specificity on DENV4) than the multiplex format (72.7% sensitivity and 58.8% specificity on DENV3; 69.6% sensitivity and 58.8% specificity on DENV4).
For IgM detection, the traditional format had lower sensitivity but higher specificity (69.6% sensitivity and 85.2% specificity on DENV3; 44.4% sensitivity and 77.4% specificity on DENV4) than the multiplex format (82.6% sensitivity and 59.3% specificity on DENV3; 66.7% sensitivity and 64.5% specificity on DENV4) (Table 2a & 2b). Cross-reactivity was observed in all three dengue rapid tests when Zika clinical samples were used for evaluation. One of the five Zika PCR positive samples tested positive (20%) on all three rapid tests. Higher cross reactivity was observed in 15 anti- Zika antibody positive samples with the lowest percentage of 47% (7 out of 15) on the InBios traditional format, 67% (10 out of 15) on the SD Dengue Duo, and the highest percentage of 73% (11 out of 15) on the InBios multiplex format.
Conclusion
No FDA-cleared rapid test is currently available for the clinical diagnosis of dengue infection in the U.S. or in a deployment setting of U.S. military personnel even though some dengue rapid tests are available overseas commercially or as research use only status (e.g., SD BIOLINE Dengue Duo). Previously, other studies showed that the SD BIOLINE Dengue Duo test had sensitivities in the range of 83.7% to 93.9% and specificities in the range of 83.9% to 98.8% [11,17- 21]. Interestingly, in this study, we found that the InBios traditional format had a similar performance to the commercially available SD BIOLINE Dengue Duo test by using a larger number of DENV3 and DENV4 samples. Upon further evaluation, we found that the InBios traditional format had a higher sensitivity and specificity than the multiplex format. The InBios traditional format also uses less sample volume than the SD Duo test (65 μl vs. 110 μl) thus making it the best candidate to be used in the multicenter clinical trial as part of the process to obtain an FDA clearance.
This study complemented and also supported our previous study that traditional format had a better performance than the multiplex format [10]. Moreover, we used an additional panel of clinical samples from Cambodia verified by different traditional diagnostic methods for dengue infection. When comparing the results for the SD Duo test, the sensitivities for the traditional format (94.8% for DENV3 and 100% for DENV4) and the multiplex format (93% for DENV3 and 86.7% for DENV4) using samples from Cambodia were very similar to the results (traditional format: 100% for both DENV3 and DENV4; multiplex format: 83.3% for DENV3 and 96.3% for DENV4) using clinical samples from other geographic regions such as Peru, Venezuela, and Thailand in our previous evaluation [10].
The additional panel of 130 samples in this study (50 DENV3 samples, 40 DENV4 samples, 10 acute non-DENV febrile samples, and 10 normal human sera along with 20 Zika clinical samples) further enhanced our confidence on the overall sensitivity and specificity to choose between the two InBios ICT prototypes. The slightly lower overall sensitivity for both formats when compared to the diagnostic results determined by traditional diagnostic assays performed at NAMRU-2 as the reference standard was because of some acute samples do not have detectable NS1. Although earlier reports showed that some ICTs had lower sensitivity on DENV4 samples [11,21], our study showed that both InBios ICTs actually had high sensitivity on DENV4 samples that may be caused by a higher sensitivity of the monoclonal antibodies from InBios to DENV4. However, the detailed design, construction, and reagents of both ICT prototypes were proprietary to InBios.
In conclusion, the InBios traditional dengue ICT is simple, easy to perform without needing sophisticated equipment, rapid, sensitive, and specific, supporting its use for a rapid and reliable diagnosis of dengue infection in resource-limiting settings. However, it should be noted that Zika positive serum samples did cross-react with this InBios traditional dengue ICT (20% and 47% for Zika PCR and antibody positive samples, respectively). Therefore, special caution needs to be taken when performing the test in an area where Zika is co-circulating or additional efforts should be placed by the company to minimize cross-reactivity with Zika samples.
Acknowledgements
This work was funded by the support agreement #IAA GT&C#N3239818GTC10338 between the Naval Medical Research Center and the U.S. Army Medical Materiel Development Activity.
Conflict of Interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author Contributions
Study concept and Design: Hua-Wei Chen and Shuenn-Jue L. Wu. Analysis and Interpretation of data: Hua-Wei Chen, Tania D. Maldonado, Christina M. Farris, Victor A. Sugiharto, Mark P. Simons, Chonthida Supaprom, and Phireak Hip. Draft of the manuscript: Hua-Wei Chen and Shuenn-Jue L. Wu. Obtained Funding: Calli M. Rooney and Shuenn-Jue L. Wu. Study supervision: Maya Williams and Gabriel N. Defang.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Army, the Department of Defense, or the U.S. government. Christina M. Farris, Maya Williams, Gabriel N. Defang, Mark Simons, Calli M. Rooney, and Shuenn-Jue L. Wu are employees of the U.S. Government. This work was prepared as part of their official duties. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties. Hua-Wei Chen, Tania D. Maldonado, and Victor A. Sugiharto are employed by the Henry M. Jackson Foundation for the Advancement of Military Medicine and are funded to do this work by the U.S. Government.
References
- Bhatt S, Gething PW, Brady OJ, Jane P Messina, Andrew W Farlow, et al. (2013) The global distribution and burden of dengue. Nature 496(7446): 504-507.
- (2019) Center for Disease Control and Prevention: Dengue fact sheet. Atlanta, GA, CDC, Georgia.
- World Health Organization: Dengue and Severe Dengue Fact Sheet. Geneva, WHO, 2020, Switzerland.
- Thomas SJ, Strickman D, Vaughn DW (2003) Dengue epidemiology: virus epidemiology, ecology, and emergence. Adv Virus Res 61: 235-289.
- US Army Medical Research and Materiel Command. The Global Ranked list of Infectious Diseases Threats. U.S. Military Infectious Disease Threats Prioritization Panel, MIDRP MEMO# MCMR-RTI, May 1, 2019.
- Burnette WN, Hoke CH, Scovill J, Kathryn Clark, Jerry Abrams, et al. (2008) Infectious diseases investment decision evaluation algorithm: a quantitative algorithm for prioritization of naturally occurring infectious disease threats to the U.S. military. Mil Med 173(2): 174-181.
- Recker M, Vannice K, Hombach J, Jit M, Simmons CP, et al. (2016) Assessing dengue vaccination impact: Model challenges and future directions. Vaccine 34(38): 4461–4465.
- Santiago GA, Vergne E, Quiles Y, Joan Cosme, Jesus Vazquez, et al. (2013) Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis 7(7): e2311.
- Namekar M, Ellis EM, O’Connell M, Joe Elm, Alexandra Gurary, et al. (2013) Evaluation of a new commercially available immunoglobulin M capture enzyme-linked immunosorbent assay for diagnosis of dengue virus infection. J Clin Microbiol 51(9): 3102-3106.
- Chen HW, Maldonado TD, Lee CR, Maya Williams, Gabriel N Defang, et al. (2021) Independent evaluation of two prototype immunochromatographic tests for dengue fever developed by InBios. Mil Med usaa553.
- Pal S, Dauner AL, Valks A, Brett M Forshey , Kanya C Long, et al. (2015) Multicountry prospective clinical evaluation of two enzyme-linked immunosorbent assays and two rapid diagnostic tests for diagnosing dengue fever. J Clin Microbiol 53(4): 1092-1102.
- Kikuti M, Cruz JS, Rodrigues MS (2019) Accuracy of the SD BIOLINE Dengue Duo for rapid point-of-care diagnosis of dengue. PLoS One 14(3): e0213301.
- Lee H, Ryu JH, Park HS, Ki Hyun Park, Hyunjoo Bae, et al. (2019) Comparison of Six Commercial Diagnostic Tests for the Detection of Dengue Virus Non-Structural-1 Antigen and IgM/IgG Antibodies. Ann Lab Med 39(6): 566-571.
- Blessmann J, Winkelmann Y, Keoviengkhone L, Vatsana Sopraseuth, Simone Kann, et al. (2020) Assessment of diagnostic and analytic performance of the SD Bioline Dengue Duo test for dengue virus (DENV) infections in an endemic area (Savannakhet province, Lao People's Democratic Republic). PLoS One 15(3): e0230337.
- Antony SJ, Chonthida S, Luy M, Hip Phireak M, Sopheab Heng, et al. (2020) A prospective study conducted among participants reporting aute febrile illness in a cohort of Periurban and rural Cambodians. Infect Dis Clin Pract 28(6): 366-370.
- Food and Drug Administration (FDA). 2016. Zika virus emergency use authorization. FDA, Washington, DC, USA.
- Tricou V, Vu HT, Quynh NV, Chau VV Nguyen, Hien T Tran, et al. (2010) Comparison of two dengue NS1 rapid tests for sensitivity, specificity and relationship to viraemia and antibody responses. BMC Infect Dis 10: 142.
- Blacksell SD, Jarman RG, Bailey MS, Ampai Tanganuchitcharnchai, Kemajittra Jenjaroen, et al. (2011) Evaluation of six commercial point-of-care tests for the diagnosis of acute dengue infections: The need for combining NS1 antigen and IgM/IgG antibody detection to achieve acceptable levels of accuracy. Clin Vaccine Immunol 18(12): 2095-2101.
- Andries AC, Duong V, Ngan C, Sivuth Ong, Rekol Huy, et al. (2012) Field evaluation and impact on clinical management of a rapid diagnostic kit that detects dengue NS1, IgM and IgG. PLoS Negl Trop Dis 6(12): e1993.
- Gan VC, Tan LK, Lye DC (2014) Diagnosing dengue at the point-of-care utility of a rapid combined diagnostic kit in Singapore. PLoS One 9: e90037.
- Osorio L, Ramirez M, Bonelo A, Luis A Villar, Beatriz Parra, et al. (2010) Comparison of the diagnostic accuracy of commercial NS1-based diagnostic tests for early dengue infection. Virol J 7: 361.




 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.