Short Communication 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Detection of Chikungunya virus in Saliva and Urine Samples of Patients from Rio de Janeiro, Brazil. A Minimally Invasive Tool for Surveillance
*Corresponding author: Monica Ferreira Moreira, Laboratório de Bioquímica e Biologia Molecular de Vetores, Departamento de Bioquímica, Instituto de Química, Caixa Postal 68563, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, 21941-909, Brasil
Received: February 25, 2021; Published: March 08, 2021
DOI: 10.34297/AJBSR.2021.12.001728
Abstract
In this study, we collected saliva and urine samples from individuals in the metropolitan region of Rio de Janeiro, Brazil, during the years of 2017 through 2019 and we were able to detect the presence of Chikungunya virus genome in these samples. Our findings reinforce the possibility to monitor Chikungunya virus circulation by analyzing saliva and urine from individuals during inter-epidemic periods.
Keywords: Chikungunya virus, Saliva and urine samples, Epidemiological surveillance of arbovirus, SYBR Green assay.
Abbreviations: CHIKV: Chikungunya Virus; qPCR: Quantitative Polymerase Chain Reaction; RT-PCR: Reverse Transcription-Polymerase Chain Reaction; ML: Maximum Likelihood
Introduction
Chikungunya virus (CHIKV) is a mosquito-borne virus, transmitted in the urban area mainly by the Aedes aegypti and Aedes albopictus mosquitoes [1]. CHIKV belongs to family Togaviridae and genus Alphavirus and consists of a single-stranded RNA genome virus, about 70 nm diameter and a phospholipid envelope [2]. This virus was first described in Africa (Tanzania) in 1954 and later identified in Asia, and it was responsible for outbreaks in these two continents from the 1960s to the 1980s [3]. Currently, CHIKV is considered a real threat to countries localized in temperate and tropical zones that are infested by Aedes spp, such as Europe and the Americas [3,4].
Human CHIKV infection results in a spectrum of manifestations and it begins with a silent incubation period lasting 2-4 days on average (range 1–12 days) [5], which can evolve to either an asymptomatic and subclinical outcome or to clinical manifestations. Clinical onset is abrupt, with high fever, headache, back pain, myalgia, and arthralgia; the latter can be intense, affecting mainly the extremities (ankles, wrists, phalanges) but also large joints, referred to as the effect of the incapacitating arthralgia [6].
Our group collected saliva and urine from volunteers presenting symptoms compatible with the disease, except for one healthy individual (male) from which saliva was collected. The individuals that had saliva collected were one male and four females. The individuals that had urine collected were two males and four females. Three females donated both, saliva and urine. The samples were collected from individuals at the Federal University of Rio de Janeiro, using approved Protocol Ethics: 80709 HUCFF/FM/ UFRJ from 2017 through 2019. Volunteers were students, student relatives and staff from the University Campus, and the ages were between 25 and 45 years old.
The saliva samples (n=5) were obtained from five individuals with arbovirus-like symptoms and one asymptomatic individual. In the urine group (n=6) all individuals were exhibiting arboviruslike symptoms. The total number of samples, therefore, was eleven (n=11) and the total number of individuals was eight (n=8). Individual samples were submitted to RNA extraction by the TRIzol™ method Reagent (Invitrogen, Carlsbad, California, USA), followed by a Reverse Transcriptase (RT) assay by superscript IV (Invitrogen, Carlsbad, California, USA), following the manufacturer recommendation. CHIKV RNA detection was performed by using RT 2-step-qPCR and conventional PCR, using the forward 5’-ctttggagccaacgctatcgctt-3’ (SGCK-F) and reverse 5’-tttgtccttgcactctgctgta-3’ (SGCK-R) primers in the standardization SYBR Green qPCR assays. For the conventional PCR, Nested methodology was used in the first reaction with the forward 5’-taccgtataagactctagtc-3’ (Nestalpha-F) and reverse 5’-tgaatgtccccaaatcttccagg-3’ (Nestalpha-R) primers, followed by the second reaction with primers (SGCK-F/SGCK-R). The amplified viral material was visualized on a 1.5% (v/v) agarose gel (Figure 1D).
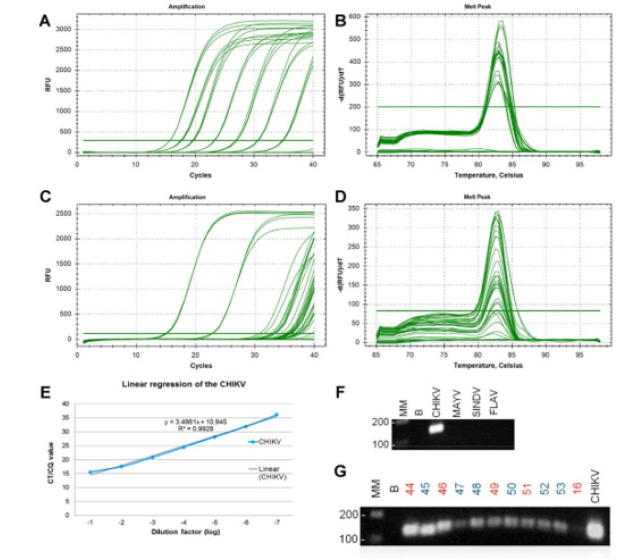
Figure 1: (A) Amplification curve for the qPCR standardization and sensitivity determination and (B) Melting pattern of the serial dilution curve for the CHIKV cDNA for the standardization with the SGCK-F / SGCK-R primers for qPCR with SYBR Green. (C) Amplification curve for the urine and saliva samples and (D) Melting pattern of the serial dilution curve for the CHIKV cDNA (positive control), saliva and urine cDNA samples for quantification with the SGCK-F / SGCK-R primers for qPCR. (E) Linear regression for the serial dilution curve with the CHIKV cDNA for standardization with the SGCK-F / SGCK-R primers in qPCR with SYBR Green, which made it possible to determine the efficiency of the reaction in the value of 93,1%. (F) qPCR gel for the cDNAs standard CHIKV cDNA with the SGCK-F / SGCKR primers; MM: molecular mass; B: blank; CHIKV, Chikungunya virus; MAYV, Mayaro virus; SINDV, Sindbis virus; FLAV, pool of flavivirus (DENV1-4, YFV, ZIKV). (G) qPCR gel for the CHIKV cDNAs (saliva and urine) and standard CHIKV cDNA with the SGCK-F / SGCKR. primers; MM: molecular mass; B: blank; red: saliva; blue: urine samples
The standardization of the SYBR GREEN qPCR assay showed a efficiency of 93.1% and specific for the detection of CHIKV genetic material as shown in (Figure 1A & 1B) (Table 1). The qPCR results showed that all urine samples were positive for CHIKV RNA, including the sample from the asymptomatic individual. Four saliva samples were positive, and one was negative. The negative saliva sample was from the donor that had arbovirus-like symptoms but donated saliva only. All two individuals that had symptoms and donated both saliva and urine had both samples positive. Overall, we had ten positive samples for CHIKV and one negative (Figure 1E) (Table 1). The generic RT-PCR for alphaviruses showed positive results, and direct sequencing of the viral amplicons showed a CHIKV-specific sequence. The result of the Sanger sequencing was used as query for BLAST searches of NCBI.
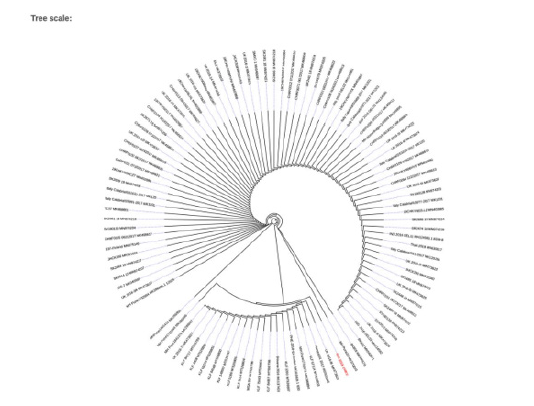
Figure 2: Phylogenetic tree using the sequences searched in BLASTn as dataset to alignment in MUSCLE software and using IQTREE to the construction of the phylogenetic tree, highlight in red the RIO2019CHIKV genome sequenced this work.
In order to identify query sequences were used to search sequences on the CHIKV genome on the NCBI genome database, using BLASTn [7]. Sequences resulting from these searches, showing more than 90% identity were deemed as homologous. All the sequences from the homologous were then used as Datasets to the aligned using the MUSCLE software [8] and then pruned for removal of regions with a high frequency of indels using TrimAL using the “-gappyout” comman [9].
The maximum likelihood (ML) tree topology was inferred with the Iq-Tree 1.6 program [10]. Branch support was assessed by the ultrafast bootstrap implementation of IqTree using 1,000 replicates [11]. IqTree was executed via the command “iqtree -s infile -bb 1000”. Because no outgroup was included in our analysis, rooting of the genome CHIKV genealogy was performed using the minimal ancestor deviation method of Tria et al. [ 12]. Figure 2 shows the phylogenetic tree indicating in red the RIO_2019_CHIKV genome sequenced in the work. Our results are similar to those of Musso et. al. [13] where they reported high number of CHIKV-positive samples during a period of high CHIKV prevalence, agreeing with the epidemiological data of the metropolitan region of Rio de Janeiro at the time of collection [4,14]. In the context of endemic co-circulation of other arbovirus diseases transmitted by Ae. aegypti such as dengue and zika fevers [14,15], a fast, reliable and noninvasive diagnostic method is important for surveillance and early detection of an increase in the number of infections.
In this study, we show that it is possible to detect CHIKV in saliva and urine samples of symptomatic and asymptomatic individuals with the new primers presented by the SYBR GREEN assay. The time of sample collection in this study correlated to an outbreak in Rio de Janeiro city. Considering the endemicity of CHIKV in the state, our results strengthen literature data when the use of saliva (preferable) and urine as important tools for detecting asymptomatic individuals during inter-epidemic and pre-epidemic periods.
Ethics Approval and Consent to Participate
The Ethical Committee of RJ/BR Rio de Janeiro (80709 HUCFF/ FM/UFRJ) has approved this study.
Availability of data and material
All the supporting data generated for this manuscript and its conclusions is available in the text. No other supporting material is required.
Competing Interests
The authors declare that they have no competing interests.
Consent for publication
Written consent was obtained from the patients / family for publication.
Funding
This work was supported by the Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ-E-26/201.331/2016-REDE ZIKA), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular (INCTEM/ CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author Contributions
All authors collaborated in the study design; TSS, TESG, VGR, participated in diagnosing and managing these eight patients; TSS, ACAM, and MFM participated in epidemiological investigation; TSS, DFF, and MFM extracted and analyzed the clinical data; TSS and VGR prepared the first manuscript draft; DFF, ACAM and MFM modified the manuscript subsequently; all authors have reviewed and approved the final manuscript.
Acknowledgments
The authors are grateful to the Program for Technological Development in Tools for Health-RPT-FIOCRUZ for use of its facilities.
References
- Sergon K, Njuguna C, Kalani R, Ofula V, Onyango C, et al. (2008) Seroprevalence of Chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. American Journal of Tropical Medicine and Hygiene 78(2): 333-337.
- Strauss JH, Strauss EG (1994) The alphaviruses: gene expression, replication, and evolution. Microbiological Reviews 58: 491-562.
- Simon F, Savini H, Parola P (2008) Chikungunya: A Paradigm of Emergence and Globalization of Vector-Borne Diseases. Medical Clin North Am 92(6): 1323-1346.
- Maljkovic Berry I, Rutvisuttinunt W, Sippy R, Beltran-Ayala E, Figueroa K, et al. (2020) The origins of dengue and chikungunya viruses in Ecuador following increased migration from Venezuela and Colombia. BMC Evolutionary Biology 20: 31.
- Pialoux G, Gaüzère BA, Jauréguiberry S, Strobel M (2007) Chikungunya, an epidemic arbovirosis. Lancet Infectious Diseases 7(5): 319-327.
- Enserink M (2006) Massive Outbreak Draws Fresh Attention to Little-Known Virus. Science 311: 1085.
- Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, et al. (2008) Database indexing for production MegaBLAST searches. Bioinformatics (Oxford, England) 24: 1757-1764.
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research 32(5): 1792-179
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15): 1972-1973.
- Trifinopoulos, J, Nguyen, L.-T, von Haeseler, A, Minh, BQ (2016) W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44(W1): W232-W235.
- Hoang, DT, Chernomor, O, von Haeseler, A, Minh BQ, Vinh LS (2018) UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol 35(2): 518-522.
- Tria FDK, Landan G, Dagan T (2017) Phylogenetic rooting using minimal ancestor deviation. Nature ecology & evolution 1: 193.
- Musso D, Teissier A, Rouault E, Teururai S, de Pina JJ, Nhan TX (2016) Detection of chikungunya virus in saliva and urine. Virology Journal 13: 1-4.
- Ministério da Saúde (2019) Monitoramento dos casos de Arboviroses urbanas transmitidas pelo Aedes (dengue, chikungunya e Zika). Boletim Epidemiológico Arboviroses 51: 1-13.
- Salles TS, da Encarnação Sá-Guimarães, T, de Alvarenga, ESL, Guimarães-Ribeiro V, de Meneses, MDF, et al. (2018) History, epidemiology and diagnostics of dengue in the American and Brazilian contexts: a review. Parasites & Vectors 11(1): 264.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.