Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Methylation Patterns in the 5’ Flanking Region of The Hoxb9 Gene in Sea Turtle Embryos with Congenital Malformations
*Corresponding author: Rodolfo Martín-del-Campo, Department of Oral Health Sciences, Faculty of Dentistry, Life Sciences Institute, University of British Columbia, 2350 Health Sciences Mall, Vancouver, Canada.
Received: March 03, 2021; Published: April 28, 2021
DOI: 10.34297/AJBSR.2021.12.001783
Abstract
Sea turtles are wild animals whose embryonic development largely depends on environmental factors. The shell of sea turtles is a novelty among vertebrates and follows a Hox code and axial formula for the group. The hoxb9 gene is a transcription factor involved in the regulation of embryonic development, mainly during neural tube development and plays an important role in shaping the thoracic-lumbar transition of sea turtles. Although it is known that Hox genes are regulated by chromatin modulating proteins, DNA methylation in CpG islands may also contribute to gene regulation. Previous studies have shown a high prevalence of congenital malformations in the olive ridley sea turtle (Lepidochelys olivacea), including Schistosomus Reflexus (SR) syndrome. Here we studied the methylation profiles in the 5’ flanking region of hoxb9 comparing normal and abnormal sea turtle embryos. Our results indicate that methylation of the putative promoter region of hoxb9 in sea turtle embryos occurred, however methylated cytosines showed individual patterns, not directly related to the development of congenital malformations.
Keywords: Congenital Malformations, Locus-Specific DNA Methylation, Homeotic Genes, Hoxb9 Putative Promoter, Epigenetics, Sea Turtles, Olive Ridley Sea Turtle, Hox Proteins, Chromosomal, Homeodomain, Morphogenesis, Amplified Fragments
Introduction
Homeotic genes act as master regulators for the specification of body plans in development, regulating other genes and programming certain developmental pathways [1]. Hoxi> genes encode transcription factors sharing a conserved DNA-binding domain, known as the homeodomain [2,3], regulating cell fate and vertebral identity during embryonic development [4]. The different Hoxi> proteins are expressed in distinct, often overlapping, domains along the antero-posterior body axis of animal embryos [5]; therefore, these spatially constrained and overlapping expression domains represent a combinatorial code to generate regional diversity [6]. Interestingly, the reptile group presents a variety of shapes, and a complete inventory of Hoxi> genes in crocodiles, turtles, snakes, and lizards have been reported [7]. Turtles have the most unusual body plan among amniotes, with a dorsal shell that fuses with the ribs [8]; for this group, as well as for other tetrapods, including mammals, birds, and amphibians, a total of 39 Hoxi> genes have been reported, divided into four paralog groups (A, B, C and D) [7].
According to Böhmer & Werneburg [9], the Hoxi> code in turtles is characterized by changes in spatial expression in the vertebral column compared to the closest phylogenetic group, the archosaurs (which includes crocodiles and birds), to give rise to the appearance of the unique shell of the turtle [8,9]. The Hoxb9 gene is involved in the regulation of embryonic development [10]. In mice, the homozygous elimination of the Hoxb1-Hoxb9 cluster causes abnormalities in the cervico-thoracic spine and defects in the sternum morphogenesis [11]. In humans, deletions in the chromosomal region encompassing the HOXB1-HOXB9 cluster are associated with developmental defects, including tracheo-esophageal abnormalities [12]. Also, in humans, de novo translocation has been reported with a breakpoint near the 5’ end of the HOXB cluster in a patient with developmental delay and skeletal malformations [13]. Embryonic development is epigenetically regulated and Hoxi> genes are no exception [14,15]. The transcriptional state of Hoxi> genes is maintained by two groups of chromatins modulating proteins, acting in opposite directions: the Trithorax Group (TrxG) proteins, which activates transcription, and the Polycomb Group (PcG), which represses transcription [15- 17].
The role of DNA methylation as a transcriptional regulator has been associated to the promoters of genes containing high density of CpG sites, called CpG islands (CGIs), which remain hypomethylated even if the locus is transcriptionally silent [18]. In Hoxi> genes, CpG islands in promoters may contribute to gene regulation in cis [19]; in addition, CpG inter or intragenic islands are more susceptible to methylation, and present tissue-specific methylation in these same loci [19]. Bárcenas-Ibarra et al. [20,21] reported the presence of congenital malformations and Schistosomus Reflexus (SR) syndrome in the olive ridley sea turtle (Lepidochelys olivacea), a species considered vulnerable according to the IUCN Red List of Threatened Species [22]. This syndrome was identified in 31% of the embryos that showed congenital malformations, and its main characteristics are the lack of closure in the abdominal wall and alterations in the spinal cord [21]. Thus, the methylation profiles in the 5’ flanking region of the hoxb9 gene were compared between embryos with multiple malformations, including SR, and their normal counterparts.
Materials and Methods
Sample collection
A previous study was conducted at El Verde Camacho nesting beach in Mexico (23° 44’ 22” N, 106° 58’ 27” W) [20]. From this study, a catalog of congenital malformations was generated [20], tissue was sampled from the back of the neck and preserved in absolute ethanol at -20°C to perform the genetic analysis. Genomic DNA was extracted by the salting-out method [23] and stored at -20°C until use.
Locus-specific DNA methylation analysis
Isolation and characterization of the 5’ flanking region of the hoxb9 gene: The 5’ flanking region of the hoxb9 gene for the olive ridley sea turtle was isolated by PCR amplification. Primers were designed according to the coding region of the hoxb9 gene (HOXB9-201 ENSPSIT00000007772.1) reported for the Chinese soft-shelled turtle (Pelodiscus sinensis) identified on ENSEMBL database, and the first exon in the draft genome of the green sea turtle (Chelonia mydas) identified in the National Center for Biotechnology Information (NCBI) database; the sequences of both species were compared and primers were designed to amplify a fragment of about 948 bp (Table 1); the reverse primer was designed within the coding region of the hoxb9 gene, whereas the forward primer was designed in the putative promoter region. Amplified fragments were purified and sequenced in Macrogen Inc. (South Korea). Once the sequences were obtained, they were manually edited, and a BLAST analysis was performed against the genome of C. mydas to corroborate identity. Once the promoter sequence of the hoxb9 gene was obtained, CpG Islands (CGI) were identified with the Methprimer software [24], Transcription Factor Binding Sites (TFBS) were identified using the GPMiner software [25], which contains 4206 known binding sites that match upstream regions of human, mouse, rat, chimpanzee, and dog genes, and the Transcription Start Site (TSS) was identified with the TSSG software [26].
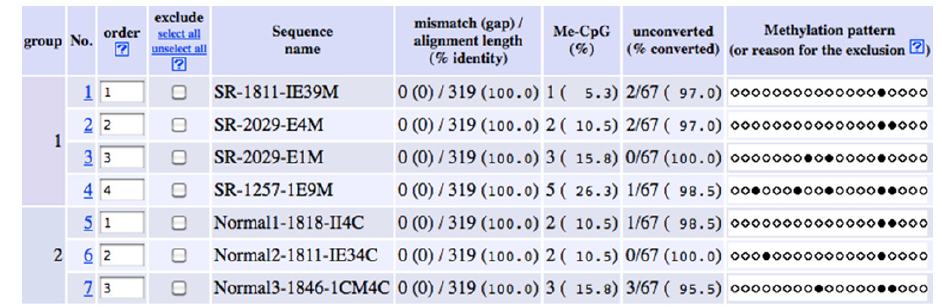
Bisulfite conversion, amplification, cloning, and analysis of methylated CpG sites: For locus-specific DNA methylation analysis, four embryos were selected that presented multiple malformations, including Schistosomus Reflexus (SR): 1) SR-1811- IE39M, 2) SR-2029-E4M, 3) SR-2029-E1M, 4) SR-1257-IE9M and 3 normal embryos with no external malformations (Table 2). The DNA of normal and SR embryos was converted with sodium bisulfite using the EZ DNA Methylation Gold Kit™ (Zymo Research) following the manufacturer’s instructions. Since CGI-1 and CGI-2 were very close, primers were designed for a single fragment of about 334bp (Table 1). PCR products were gel-purified with the Wizard® SV Gel and PCR Clean-up System (Promega). Purified PCR products were ligated into a pGEM-T cloning vector (Promega). Transformation was performed by heat shock using Escherichia coli JM109 competent cells (Promega) and plasmid extraction was performed by alkaline lysis. Six to ten clones per embryo were sequenced using the T7 universal primer, and the difficult sequencing service provided by Macrogen Inc (Korea). The frequency of methylated CpG sites was examined using the QUMA (Quantification Tool for Methylation Analysis) software: (http://quma.cdb.riken.jp).
Results
Isolation and characterization of the 5’-flanking regions of the hoxb9 gene
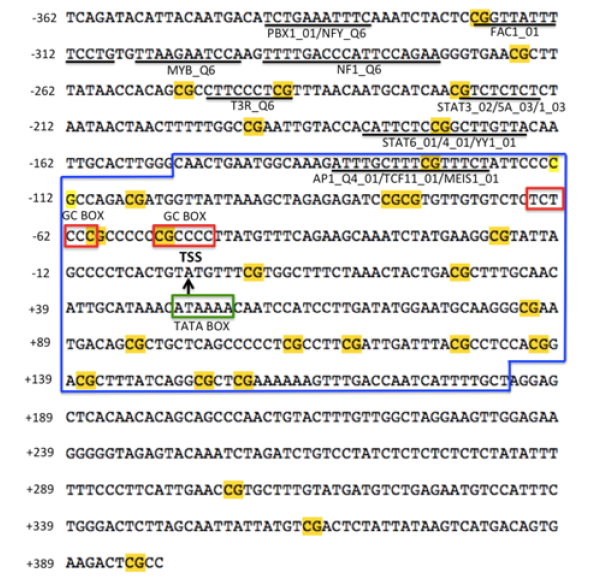
A fragment of 760 bp was obtained from genomic DNA of the olive ridley sea turtle. BLAST analysis indicated a >90% identity with the 5’-flanking regions (called putative promoter region) of the hoxb9 gene of the green sea turtle (C. mydas) and the chinese soft shelled turtle (P. sinensis). Nucleotide 363 (of the isolated fragment) was identified as a TSS (position +1) (Figure 1). The putative TATA box was located at nt +50 relative to the TSS. Fifteen putative transcription factor binding sites (TFBSs) were also identified: Pre-B-cell leukemia transcription factor 1 (PBX1_01), Nuclear transcription factor Y (NFY_Q6), Fetal Alz-50-Reactive Clone 1 (FAC1_01), MYB transcriptional activator (MYB_Q6), Neurofibromin 1 (NF1_Q6), Thyroid hormone receptor T3R (T3R_ Q6), Signal transducer and activator of transcription 3 (STAT3_02), Signal transducer and activator of transcription 5A (STAT5A_03), Signal transducer and activator of transcription 1 (STAT1_03), Signal transducer and activator of transcription 6 (STAT6_01), Signal transducer and activator of transcription 4 (STAT4_01), transcriptional repressor YY1 (YY1_01), Activator protein 1 (AP1_ Q4_01), Nuclear factor TCF11 (TCF11_01), Homeobox protein MEIS1 (MEIS1_01). Putative GC boxes were also found upstream the TSS (Figure 1).
Identification of CpG islands (CGIs) in the 5’-flanking region of the hoxb9 gene
Two CGIs were identified, CGI-1 of 109 bp (nt -120 to -12) and CGI-2 of 127 bp (nt +57 to +183) (Figure 2). A set of PCR primers (including CGI-1 and CGI-2) were designed to amplify bisulfitetreated DNA (Table 1). Primers amplified a fragment of 334 bp, which included 19 CpG sites, seven of which were within the CGI-1, and nine of them were in the CGI-2 (Figure 1 & 2). Characterization of the putative promoter region of the hoxb9 gene. CpG sites with methylation potential (yellow), possible transcription factor-binding sites (underlined in black), transcription start site (TSS, black arrow), TATA box (green rectangle) and GC box (red rectangles) are indicated. The blue box indicates the 334 bp fragment which includes the CpG island-1 (CGI-1; nt -120 to -12) and the CpG island-2 (CGI-2; nt +57 to +183). Regulatory features were identified by the GPMiner software [25]. Identification of CpG islands (light blue) in the putative promoter region of the hoxb9 gene of L. olivacea. A TSS was identified at nt 363 of the isolated fragment. CGI-1 was identified upstream of the TSS and CGI-2 in the 5 ‘UTR. CGIs were identified with the Methprimer software [24].
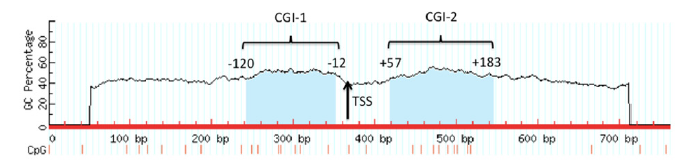
Figure 2: Identification of CpG islands (light blue) in the putative promoter region of the hoxb9 gene of L. olivacea.
Locus-specific DNA methylation in the putative promoter region of the hoxb9 gene
Bisulfite sequences from abnormal and normal embryos were compared. Conversion efficiency was 95-100% (Figure 3). Mosacism methylation was present in most cases (Table 3). A sequence of 319 bp was analyzed, which contained nineteen CpG sites Figure 3. Because most of the TFBS were identified upstream of both CGIs analyzed with bisulfite-PCR (Figure 1), it was not possible to identify methylated CpG sites in putative TFBS. All embryos analyzed (normal and abnormal) showed a particular methylation profile (Figure 3). Interestingly, there was a consistent pattern in which sites +128 and +136, located in the 5’ UTR region Figure 1 and 3 were methylated regardless of whether the embryos presented developmental defects, specifically site +128 was methylated in all embryos analyzed, which could suggest an important role of locus-specific methylation in the 5’ UTR of the hoxb9 gene related to embryonic development, but not related to any specific malformation. A different, individual pattern of methylated cytosines was also observed in the embryos, however, with such a small sample size and the variety of malformations observed is not possible to know if these methylation patterns are related to malformations or other developmental traits. Methylated cytosines were also identified in the non-CpG (CHG/CHH) context in both normal and abnormal embryos (Table 1). The role of non- CpG methylation in non-mammalian vertebrates is less understood and requires further research [27].
Discussion
An apparent heterogeneous and individual methylation profile was observed in CGIs of the putative promoter region of the hoxb9 gene in both normal and malformed embryos. Bisulfite conversion efficiency of DNA was high (95 to 100%) but mosaicism was evident (Table 3). This may be due to the sampled region (the back of the neck) since it originates from ectoderm and mesoderm derivatives [28]. It is known that developmental genes regularly remain hypomethylated even if the locus is transcriptionally silent, although some methylation could be present [19], and this is consistent with our results in both normal and malformed embryos. Although heterogeneous, the methylation profile observed in the putative promoter region of hoxb9 shows a consistent pattern in two CpG sites (+128 and +136) that are repeated in normal and abnormal embryos (Figure 3) these methylated cytosines could play a role in the regulation of hoxb9 during development and may not be involved in developmental defects [29].
In this sense, it has been reported that the 5’ UTR region of several genes plays an important role in alternative splicing processes [30], it would be interesting to know if there is a relationship between the presence of these methylated cytosines and mRNA processing. In Hox genes, tissue-specific methylation of CGIs is essential for development [19]. The patterns of DNA methylation reflect signals established during development, and it is known that they can be modified in adult tissues with tumor progression [29,31]. In fact, it is well established that proteins of the Polycomb group modulate Hox gene expression by histone marks (e.g., methylation of histone 3 at lysine 27-H3K27me-) [32], so that DNA methylation could promote transcription by antagonizing genes from the Polycomb group [33], repressing gene expression. For instance, in mice with skeletal defects, hypermethylation of DNA at Hox loci was associated with the elimination of the SET domain of the Mll gene of the Tritorax group (antagonist of the Polycomb group) [34].
Currently, it is well established that environmental factors during embryonic development can cause different epigenetic modifications, such as changes in DNA methylation or posttranslational modification of histones; these factors may include nutrient deficiency, presence of pollutants, or even stress [35]. In sea turtles, temperature, humidity, and gas exchange during incubation may behave as teratogenic agents [36], affecting development through subtle epigenetic changes in Hox (and other developmental) genes with phenotypic implications [34,37,38]. No link was observed between methylation patterns in the putative hox promoter and congenital malformations in the embryos analyzed in this study; hox regulates the development of the spine in the thoracic-lumbar region, which could be implicated in SR, and it would be necessary to sample and analyze tissue from that region. It is also important to consider that the Hox genes act in a combinatorial fashion, and different Hox genes are required to shape each vertebra and the axial structure. In addition, paralogous Hox genes present functional compensation [39,40] therefore, the regulatory effects of Hox genes should be studied together, according to the Hox code and axial formula of each species.
Conclusions
Hox genes are master regulators of the body plans during embryonic development in both vertebrate and invertebrate animals and are regulated (at least in part) by epigenetic mechanisms. In this work we identified that the 5’ flanking region of the hoxb9 gene in sea turtle embryos, in addition of presenting random and individual methylation patterns, also presents a consistent methylation pattern in some specific cytosines, suggesting an important role of locus-specific methylation in the 5’ UTR of the hoxb9 gene related to embryonic development, but not necessarily related to any specific malformation. Several questions remain regarding the role of DNA methylation in Hox genes regulation and function during normal and abnormal embryonic development, genomic and functional studies are required to address these questions, although in wild, protected animals like sea turtles these studies are still challenging.
Acknowledgements
The authors would like to thank all the people working at El Verde Camacho nesting beach, especially Daniel Ríos-Olmeda and Raquel Briseño-Dueñas. We also thank to the Wildlife Department of the Ministry of the Environment in Mexico (DGVS-SEMARNAT) for capture permits, and the volunteer Rogelio Iván Lozano-Guzmán for assistance in collecting and cleaning the olive ridley nests. We are especially grateful to Annelisse Bárcenas-Ibarra for collecting the samples and to Isaías Rojas-Lleonart for his assistance in the identification and description of congenital malformations in the olive ridley sea turtle. Thanks to Rubí Hernández-Cornejo, and Carmen Bolán-Mejía, for technical assistance, and the National Council for Science and Technology (CONACYT, Mexico) for the scholarship granted to RMC. Also, many thanks to Rodolfo Martin del Campo Gaytan and Virginia Flores for sponsoring this publication.
Declaration of Interest
The authors have no conflict of interest to declare.
References
- Gehring WJ, Hiromi Y (1986) Homeotic genes and the homeobox. Annu Rev Genet 20(1): 147-173.
- McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ (1984) A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308(5958): 428-433.
- Scott MP, Weiner AJ (1984) Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci USA 81(13): 4115-4119.
- Lewis EB (1978) A gene complex controlling segmentation in Drosophila. Nature 276(5688): 565-570.
- Lemons D, McGinnis W (2006) Genomic evolution of Hox gene clusters. Science 313(5795): 1918-1922.
- Hunt P, Gulisano M, Cook M, Sham MH, Faiella A, et al. (1991) A distinct Hox code for the branchial region of the vertebrate head. Nature 353(6347): 861-864.
- Liang D, Wu R, Geng J, Wang C, Zhang P (2011) A general scenario of Hox gene inventory variation among major sarcopterygian lineages. BMC Evol Biol 11: 25
- Ohya YK, Kuraku S, Kuratani S (2005) Hox code in embryos of Chinese soft‐shelled turtle Pelodiscus sinensis correlates with the evolutionary innovation in the turtle. J Exp Zool B Mol Dev Evol 304(2): 107-118.
- Böhmer C, Werneburg I (2017) Deep time perspective on turtleneck evolution: Chasing the hox code by vertebral morphology. Sci Rep 7(1): 8939.
- Nagel S, Burek C, Venturini L, Scherr M, Quentmeier H, et al. (2007) Comprehensive analysis of homeobox genes in Hodgkin lymphoma cell lines identifies dysregulated expression of HOXB9 mediated via ERK5 signaling and BMI1. Blood 109(7): 3015-3023.
- Medina-Martinez O, Bradley A, Ramírez-Solis R (2000) A large, targeted deletion of Hoxb1-Hoxb9 produces a series single-segment anterior homeotic transformation. Dev Biol 222(1): 71-83.
- Pajusalu S, Reimand T, Uibo O, Vasar M, Talvik I, et al. (2015) De novo deletion of HOXB gene cluster in a patient with failure to thrive, developmental delay, gastroesophageal reflux, and bronchiectasis. Eur J Med Genet 58(6-7): 336-340.
- Yue Y, Farcas R, Thiel G, Bommer C, Grossmann B, et al. (2007) De novo t (12; 17) (p13. 3; q21. 3) translocation with a breakpoint near the 5′ end of the HOXB gene cluster in a patient with developmental delay and skeletal malformations. Eur J Hum Genet 15(5): 570-577.
- Soshnikova N, Duboule D (2009) Epigenetic temporal control of mouse Hox genes in vivo. Science 324(5932): 1320-1323.
- Barber BA, Rastegar M (2010) Epigenetic control of Hox genes during neurogenesis, development, and disease. Ann Anat 192(5): 261-274.
- Mendenhall EM, Bernstein BE (2008) Chromatin state maps: new technologies, new insights. Curr Opin Genet Dev 18(2): 109-115.
- Bernd S, Martinez A, Iovino N, Cavalli G (2011) Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol 12(12): 799-814.
- Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25(10): 1010-1022.
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, et al. (2007) Distribution, silencing potential, and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39(4): 457-466
- Bárcenas A, De Cueva, Rojas I, Abreu F, Lozano I, et al. (2015) First approximation to congenital malformation rates in embryos and hatchlings of sea turtles. Birth Defects Res A Clin Mol Teratol 103(3): 203-224.
- Bárcenas-Ibarra A, Rojas-Lleonart I, Lozano-Guzmán RI, García-Gasca A (2016) Schistosomus Reflexus Syndrome in Olive Ridley Sea Turtles (Lepidochelys olivacea). Vet Pathol 54(1): 171-177.
- Abreu-Grobois A, Plotkin P (IUCN SSC Marine Turtle Specialist Group) (2008) Lepidochelys olivacea. The IUCN Red List of Threatened Species 2008: e.T11534A3292503.
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16(3): 1215.
- Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18(11): 1427-1431.
- Lee TY, Chang WC, Hsu JBK, Chang TH, Shien DM, (2012) GPMiner: an integrated system for mining combinatorial cis-regulatory elements in mammalian gene group. BMC Genomics 13(1): S3.
- Solovyev VV, Shahmuradov IA, Salamov AA (2010) Identification of promoter regions and regulatory sites. Methods Mol Biol 674: 57-83.
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462(7271): 315-322.
- Matsuoka T, Ahlberg PE, Kessaris N, Iannarelli P, Dennehy U, et al. (2005) Neural crest origins of the neck and shoulder. Nature 436(7049): 347-355.
- Flagiello D, Poupon MF, Cillo C, Dutrillaux B, Malfoy B (1996) Relationship between DNA methylation and gene expression of the HOXB gene cluster in small cell lung cancers. FEBS letters 380(1-2): 103-107.
- Mironov AA, Fickett JW, Gelfand MS (1999) Frequent alternative splicing of human genes. Genome Res 9(12): 1288-1293.
- Gopalakrishnan S, Van Emburgh BO, Robertson KD (2008) DNA methylation in development and human disease. Mutat Res Fundam Mol Mech Mutagen 647(1-2): 30-38.
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, et al. (2002) Role of histone H3 lysine 27 methylation in polycomb group silencing. Science 298(5595): 1039-1043.
- Wu H, Coskun V, Tao J, Xie W, Ge W, et al. (2010) Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329(5990): 444-448.
- Terranova R, Agherbi H, Boned A, Meresse S, Djabali M (2006) Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci USA 103(17): 6629-6634.
- Lo CL, Zhou FC (2014) Environmental alterations of epigenetics prior to the birth. Int Rev Neurobiol 115: 1-49.
- Martín-del-Campo R, Calderón-Campuzano MF, Rojas-Lleonart I, Briseño-Dueñas R, García-Gasca A (2021) Congenital Malformations in Sea Turtles: Puzzling Interplay between Genes and Environment. Animals 11(2): 444.
- Zimm R, Bentley BP, Wyneken J, Moustakas-Verho JE (2017) Environmental causation of turtle scute anomalies in ovo and in silico. Integr Comp Biol 57(6): 1303-1311.
- Martín-del-Campo R, Sifuentes-Romero I, García-Gasca A (2019) Hox genes in reptile development, epigenetic regulation, and teratogenesis. Cytogenet Genome Res 157(1-2): 34-45.
- Greer JM, Puetz J, Thomas KR, Capecchi MR (2000) Maintenance of functional equivalence during paralogous Hox gene evolution. Nature 403(6770): 661-665.
- Holland PW (2013) Evolution of homeobox genes. Wiley Interdiscip Rev Dev Biol 2(1): 31-45.






 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.