Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Patterns of Genetic Structure and Evidence of Gene Flow between Arabian Peninsula and European Populations
*Corresponding author: Hadi Charati, Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming 650204, China/State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, China.
Received: November 26, 2020; Published: April 06, 2021
DOI: 10.34297/AJBSR.2021.12.001759
Abstract
The genetic interaction was observed between Asian and European populations. However, genetic admixtures among Eurasians, particularly between East Asians and North western Europeans have been reported but population admixture and gene flow between Arabian Peninsula and European populations have been poorly studied at the level of genome. Here, we have compared the whole-exome sequencing of 1208 individuals from the Arabian Peninsula, Africa, Europe, Caucasian, East and South Asia, to identify genetic structure and gene flow between them. We have shown that there is less differentiation between Arabian Peninsula and Italy samples than that between the Arabian Peninsula and other Europeans in this study. As well as Italy samples exhibit higher similarity of copy number variation of deletion and duplication with Qatari individuals. Arabian Peninsula ancestry expanded into the South Asian, Caucasian and parts of European populations. Large identity by descent tracts (≥ 2.0 cM) were identified between Arabian Peninsula and individuals from Kenya and Nigeria. outgroup f3-statistics suggest that within Europeans, Southern Europeans share more genetic drift with Arabian Peninsula than with other European regions. It is possible that these patterns reflect the Arabs migration to the Italian island of Sicily, perhaps dating back to the 831-1072 AD. Our results showed genetic structure of Afro-Eurasian populations, with different levels of southern European admixture, as a result of the genetic interaction with Arabian Peninsula.
Keywords:Arabian Peninsula; European populations; Gene flow; Identity by descent; Whole-exome sequencing
Introduction
The genetic structure of biological populations varies across the world, as results from the interaction between population admixture, movement, gene flow and natural selection. It is believed that early modern humans left Africa via the Nile Valley heading to the Middle East and through the Red Sea crossing to the Arabian Peninsula (AP) and settling in these places as early as 125,000 years ago [1,2]. Then ancestors began to spread into Southern Asia and Australia [3], Europe, and eventually, the Americas [4] and being the basis of these modern human population structures, and a continuing path for their admixture.
Analysis of Europeans ancient DNA (aDNA) have afforded a complex population genetic history of this regions involving at least three major migrations of prehistoric people that were influenced by Environmental conditions [5]. Higher level of genetic diversity are observed in southern Europe compared with other regions of the continent as results of gene flow across the southern Europe over the thousand years ago [6,7]. However, the analysis of aDNA from South eastern Europe has revealed the presence of the Caucasus ancestral [8] and suggests a complex network of ancient ancestry of these regions [9].
Genetic interactions have been reported between Asian and European ethnic groups. These interactions has been affected by many factors, such as the impact of great empires (Roman and Mongolian empires) and the ancient Silk Route (206 BC) as communication highway between European and Central Asian populations. It has been suggested that most genetic admixtures observed between 310 and 2,400 years ago with the very low-level admixture in North western European and East Asian populations and the highest admixture events in Central Asian populations [10]. In addition, genome-wide studies revealed that European Romani individuals fall between Southern Asian and non-Roma European populations relative to populations from current Punjab state of India, Central Asia, Pakistan and Caucasus [11-13]. Furthermore, Gene flow from North Africa groups affected the gene pool of differential human populations in southern Europe [6].
However, genetic interaction among Eurasians groups, particularly between North Europeans and East Asians has been reported [14,15], the AP gene flow in Europeans has not been well studied yet. To overcome the mentioned limitations, the present research consists of a whole-exome sequencing (WES) analysis of the 1208 individuals from the Arabian Peninsula, Africa, Europe, Caucasian, East and South Asia, with the following aims to determine the level of admixture of the AP with other Europeans and to distinguish the patterns of gene flow between them. Our main objectives were to evaluate the status of genetic structure, phylogenetic relationships and quantify the extent and pattern of recent gene flow between AP and European populations.
Methods
Study samples and variant calling
The Illumina WES of AP (Saudi Arabia and Qatar; 298 samples), Europe (British, Germany, Italy, Northern and Western Europe (NW Europe); 195 samples), Africa (Nigeria and Kenya; 164samples), Caucasian (Armenia, Azerbaijan, Georgia; 27 samples), East Asia (China, Korea and Japan; 319 samples), and South Asia (Pakistan, India and Sri Lanka; 205 samples) were downloaded from the Sequence Read Archive (SRA) at the NCBI site (https://trace.ncbi. nlm.nih.gov/Traces/sra) (Table S1 & S2). Briefly, raw sequences were aligned to the human reference genome (version hg 19) using Burrows-Wheeler Aligner (BWA) (https://sourceforge.net/ projects/bio-bwa) [16]. All SAM files were converted to the BAM files, Using SAM tools [17], followed by sorting and indexing. PCR duplicates were marked from the BAM files using Mark Duplicates tool from Picard Tools (https://github.com/broadinstitute/ picard). By using Indel Realigner and Base Recalibrator command from GATK 3.4 program, indels were realigned and base quality was recalibrated respectively (www.broadinstitute.org/gatk). Finally, the SNPs related to all individuals were detected and filtered using the Unified Genotyper with the “EMIT-ALL-SITES” option and the Variant Filtration command in the GATK 3.4 program. VCF file was used to a filtering for MAF ≥0.05 and max-missing = 0.90 by using VCFtools [18] v.0.1.13. After the application of quality control filters, 203,256 high-quality WES SNVs were retained for our analyses.
Mixture analysis
To investigate the potential of genetic admixture between AP, European and worldwide population samples, we have used the block relaxation algorithm implemented in ADMIXTURE [19]. v.1.3.0 to estimate individual ancestry proportions given k ancestral components. We have applied the default cross validation parameters (folds = 5) with itera¬tions of k value ranging from 2 to 23. Minimum squared error values calculated from the crossvalidation procedure in ADMIXTURE to evaluate the fit of different values of k determined that k = 12 was optimum for samples (Figure S1).
Copy number variations (CNVs) analysis
CNVs were recognized using CNVkit [20], a command-line toolkit, to visualize and infer copy number from WES data to a reference human genome (hg19). For this purpose, 5 samples from each population were randomly selected to show more details in the heatmap. We used bam files as input and default CNVkit settings were used for CNV identification individually. Given CNVs in X and Y chromosome were not included.
Principal-components analysis (PCA)
PCA was used to investigate the affinities in human populations and the relationships between them. We have performed PCA on all samples using smart pca from the Eigen strat package [21] and the first two principal components were compared graphically.
Influence of recent migration on maximum-likelihood phylogeny
Tree mix v.1.13 [22] was used to estimate the historical relationships and migration among populations on the maximumlikelihood phylogeny. We have tested the fit of model 3 migration events (-m 3). Tree Mix was run using SNPs grouped in windows of 500 (--k 500) with samples grouped by location and population. Sample-size adjustment was turned off because samples per population were ≥ 7.
Gene flow
To study the degree of genetic relatedness between AP and worldwide population, we computed outgroup f3-statistics [23,24] of the form f3 (Mbuti; AP,X). This statistic measures the level of shared genetic drift between AP and population X, after divergence of the ancestors of Mbuti as outgroup. Standard error was estimated using a weighted block jack knife approach [25] over 5-Mb blocks. Regions of genomic identity by descent (IBD) were identified [26] with the Beagle [27] implementation of fastIBD [28].
Results
Genome-wide ancestry analysis of the AP and worldwide population
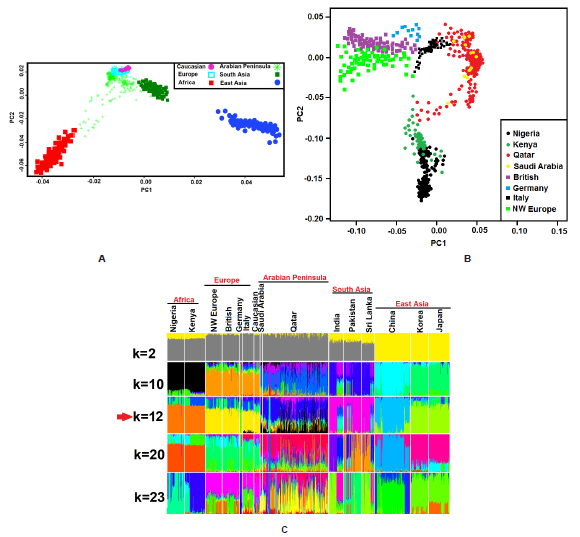
Figure 1: Population structure analysis. Principal component analysis (PCA) with samples from the (A) worldwide population (B) AP, African and European populations. (C) ADMIXTURE analysis; the lowest cross-validation error was observed at K=12.
We have applied PCA and the clustering algorithm ADMIXTURE to study the population structure and relationship of AP to European and worldwide population in a WES of 1208 samples. In Figure 1A, we have tested the relationships among worldwide population. AP fall between the African, Europeans and South Asian populations, consistent with AP ancestry derived from African, European and South Asian populations (Figure 1A & 1C). Population of the European and Caucasian were found less separated and located in a top position in the PCA plot. East Asians show the most distance from AP populations compared with non-East Asians groups (Figure 1A). In Figure 1B, we have repeated the PCA using only the European, African and AP populations to show more details of the genetic structure. We have observed that there is less differentiation between AP and Italy samples than that between the AP and other Europeans. Furthermore, AP populations showed genetic diversity and wide spread on the PC2 axis and tend to show a greater affinity with Kenya samples (Figure 1B). The long tails exhibited by the AP populations in the PCA plot resulting admixture events or the event of gene flow from other populations (Figure 1B).
When performing the ADMIXTURE analysis, the lowest crossvalidation error could be found when K=12 (Figure S1). These results clearly show that the AP ancestral patterns expansion into the South Asian, Caucasian, parts of European, especially Italy individual, which were consistent with the observed PCA results and suggested admixture or the occurrence of gene flow events (Figure 1C). All AP subgroups showed similar ancestral patterns. However, we have observed small portions of other ancestral components in AP individuals (Figure 1C).
To test signatures of recent admixture of modern human populations in this study, we have created a maximum likelihood tree using the Tree Mix approach [29]. Tree Mix uses a model that allows for both population splits and gene flow to better capture historical relationships between populations. We first generated a tree with no migration events (Figure 2A). The evolutionary history of localities without migration showed a close relationship among the AP population that confirmed our PCA and ADMICTURE result. Furthermore, the tree shows that the European and Caucasian shared drift with AP populations (Figure 2A). Italy individuals was closed the AP branch but showed substantial divergence. Germany individuals showed much greater apparent divergence among subjects from European populations (Figure 2A). Allowing three admixture events finds evidence of admixture between African and the AP and the Pakistan populations, and between the Caucasian and the Germany and East Asian populations (Figure 2B).
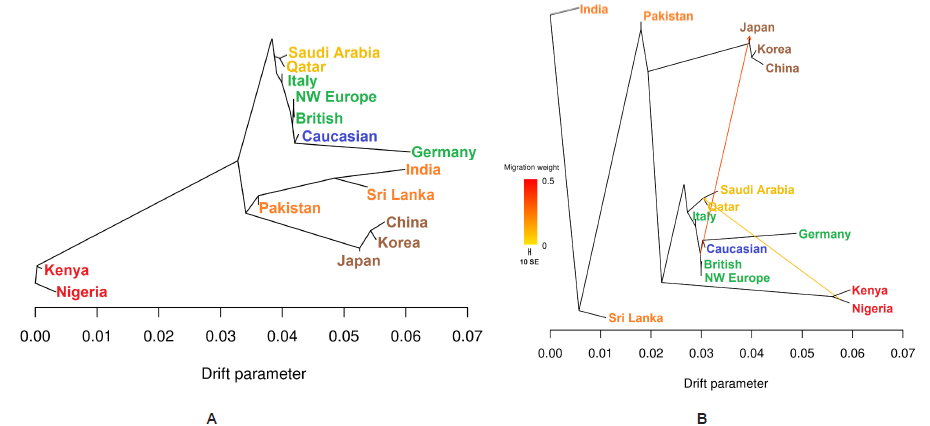
Figure 2: (A) Tree Mix tree with no migration events (B) Tree Mix tree with three migration event. The graph is stable in 100 bootstrap replicates. Migration arrows are coloured according to their weight.
Whole-Exome CNV analysis of AP and worldwide population
CNVs are a category of structural variation determined by the gain or loss of large regions of genomic sequence [30,31] and can be used to measure genetic relatedness. In the present study, we have explored the structure of CNVs, based on the CNV count per individual across AP and worldwide population. We have performed a heat map analysis using the top 5% of CNVR variances (we have considered only CNVR contain at least CNV>2 events), and we have observed the consistent patterns based on CNV count. The heat map based on frequency of CNV events clearly arranged groups according to the geographic origin of populations (Figure 3). Focusing on the heat map, we have observed that Italy exhibit higher similarity of deletion and duplication with Qatari compared to the other geographic origins (Figure 3). While the south Asians show the highest frequency of deletion events per sample we identify high deference of CNV between European and East Asian populations.
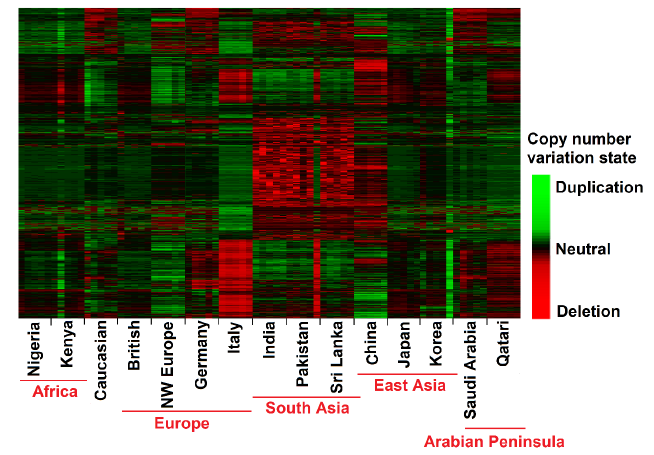
Figure 3: Copy number states of AP and worldwide population. These maps represent CNVs estimates across the human genome. The color of each window represents its copy number, as indicated by the scale on the right. The heat map was computed based on the average log ratio of corresponding probes measured in both replicates. Chromosomes and individuals are represented on the Y-axis and X-axis, respectively.
Gene flow between AP and worldwide human populations
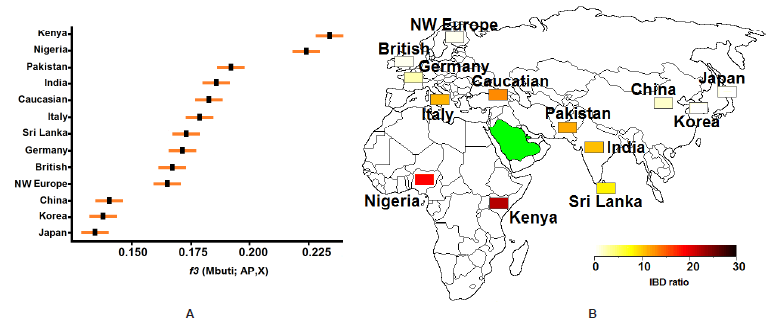
Figure 4: Gene flow between AP and worldwide human populations. (A) Statistic f3 values obtained with pop stats when taking Mbuti as outgroup. The statistic and one standard error deviation are presented for each combination test. (B) Shared IBD segments between AP and worldwide populations. The IBDs with length ≥ 2 cM and significant score < 10−5 were included. The degree of gene flow is measured as IBD sharing statistics.
To examine which populations show the highest shared genetic drift with the AP genome, we applied outgroup f3-statistics (Reich et al., 2009). Using Mbuti Pygmy as outgroup, we have found that Kenya and Nigeria have the greatest affinity (i.e. the largest f3) to AP among 13 contemporary populations, (Figure 4A & Table S3). Within Asia, The highest sharing was found with Pakistani individuals. Within Europe, we have found the highest level of shared genetic drift for Italy and, to a lesser extent, for NW European population (Figure 4A).
The use of WES data and a larger sample size in our study allows us to better investigate the level of admixture and characterize recent gene flow by exploring for long segments of genetic identity by descent (IBD). Large IBD tracts (≥ 2.0 cM) were identified between AP and both populations from Africa in this study, and the degree of recent gene flow with Eurasian populations ranged from essentially none with East Asians to very high with the Caucasian and Italy individuals (Figure 4B). These results are consistent with geographical proximity between AP and each respective group. Of the populations included in Eurasian, Pakistan shares the second-highest level of IBD with the AP people, behind Caucasian population (Figure 4B).
Discussion
Analyzing the detailed genetic diversity among different human ethnic group can be biomedically beneficial, as well as identifying stratifications within populations and interactions among populations and suggesting shared ancestry through time and across geographic regions. In the present study, WES data have revealed that recent genetic admixture did occur and have been prevalent in Europe continent [32]. Admixture has been detected between Southern European (Italy) and AP populations (Figure 1C) which are geographically far away from each other and generally considered as well-differentiated populations. We have observed that this genetic admixture might not exactly came from AP, instead, it could come from some Caucasian people [33] who live in Eastern Europe. Qatari samples were overlap with all Saudi Arabia, part of Italy, south Asian and African populations. Previous study reported that Qatari can be separated into three founder populations: Iranian (‘‘Persian’’), Arab and Central Asia and Bantu-speaking Africans [34]. According to our finding in the present study Qatari founder can be more than three and South Asian and European populations can be initial origination of the founders of Qatari (Figure 1C).
Differences in copy number of genomic segments can result in changes in gene expression and phenotypic variation through gene disruption and altering gene dosage [35,36]. Based on CNV analysis, our study revealed CNV structure similarity between Southern Europe (Italy) and AP populations [37]. Previous study of Y-chromosome has highlighted examples of male- mediated gene flow from AP to European populations [38]. Here, we showed that gene flow from AP to European is not merely reflected on the Y chromosome but corresponds to a much broader effect and have likely had an effect on genetic diversity between southern and northern European populations. We have found that NW Europeans, British and Germany populations did not show levels of admixture with AP. These findings are consistently reconstructed by different methods including our Tree Mix, ADMIXTURE and PC analyses.
We detect high levels of IBD and genetic drift between the AP and the majority of the surrounding populations. In European populations, both IBD and f3-ratio with the AP appear elevated in southern Europe (Italy). It is possible that these patterns reflect the Arabs migration to the Italian island of Sicily, perhaps dating back to the 831-1072 AD [39]. In addition, Caucasian individuals are closely related to AP populations, in agreement with a continuous gene flow, as clearly determined with IBD and f3-ratio statistics [40,41]. As well as AP is similar to African populations, this finding confirms that the Red Sea coasts may have been important in this southern expansion [42]. IBD analysis revealed that the Kenya and Nigeria exhibit the highest IBD sharing with the AP (≥ 2 cM). This may suggest not only recent gene flow between populations, but also their common ancestry or ancient admixture.
Conclusion
Our findings contribute to an improved understanding of the history of human migration and the evolutionary mechanisms that have shaped the genetic structure of populations in Afro- Eurasia. Our study has confirmed that the southern European, Southern Asian and Caucasian populations have ancestry from AP. Furthermore, the extreme similarity between AP and Southern European (Italy) population provides evidence of significant gene flow between these two regions. This information will be useful in further investigations of human population movements and possibility of gene flow in other regions of the world.
Acknowledgment
Thanks are owed to the University of Tabriz Research Computing support staff for analytic assistance.
Conflict of interest
The author(s) declare that there are no competing interests.
Author Contributions
HC and RJO performed data analysis, manuscript preparation and manuscript revision. Both the authors have read and approved the final manuscript.
Data Availability Statement
Data are available in the Supporting Information.
References
- Armitage SJ, Jasim SA, Marks AE, Parker AG, Usik VI, et al. (2011) The southern route “out of Africa”: Evidence for an early expansion of modern humans into Arabia. Science 331(6016): 453-456.
- Lawler A (2011) Did Modern Humans Travel Out of Africa Via Arabia? Science 331(6016): 387-387.
- Rasmussen M, Guo X, Wang Y, Lohmueller KE, Rasmussen S, et al. (2011) An Aboriginal Australian genome reveals separate human dispersals into Asia. Science 334(6052): 94-98.
- López S, Van Dorp L, Hellenthal G (2016) Human Dispersal Out of Africa: A Lasting Debate. Evolutionary Bioinformatics 11(Suppl 2): 57-68.
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, et al. (2014) Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513(7518): 409-413.
- Botigue LR, Henn BM, Gravel S, Maples BK, Gignoux CR, et al. (2013) Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proceedings of the National Academy of Sciences 110(29): 11791-11796.
- Currat M, Excoffier L (2005) The effect of the Neolithic expansion on European molecular diversity. Proceedings of the Royal Society B: Biological Sciences 272(1564): 679-688.
- Lazaridis I, Mittnik A, Patterson N, Mallick S, Rohland N, et al. (2017) Genetic origins of the Minoans and Mycenaeans. Nature 548(7666): 214-218.
- Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, et al. (2016) Genomic insights into the origin of farming in the ancient Near East. Nature 536(7617): 419-424.
- Qin P, Zhou Y, Lou H, Lu D, Yang X, et al. (2015) Quantitating and Dating Recent Gene Flow between European and East Asian Populations. Scientific Reports 5: 9500.
- Font-Porterias N, Arauna LR, Poveda A, Bianco E, Rebato E, et al. (2019) European Roma groups show complex West Eurasian admixture footprints and a common South Asian genetic origin. PLoS Genetics 15(9): e1008417.
- Mendizabal I, Lao O, Marigorta UM, Wollstein A, Gusmão L, et al. (2012) Reconstructing the population history of European Romani from genome-wide data. Current Biology 22(24): 2342-1349.
- Moorjani P, Patterson N, Loh PR, Lipson M, Kisfali P, et al. (2013) Reconstructing Roma history from genome-wide data. PLoS One 8(3): e58633.
- Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, et al. (2014) A genetic atlas of human admixture history. Science 343(6175): 747-751.
- Lipson M, Loh PR, Levin A, Reich D, Patterson N, et al. (2013) Efficient moment-based inference of admixture parameters and sources of gene flow. Molecular Biology and Evolution 30(8): 1788-1802.
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25(14): 1754-1760.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) Genome Project Data Processing Subgroup, The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16): 2078-2079.
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, et al. (2011) The variant call format and VCFtools. Bioinformatics 27(15): 2156-2158.
- Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Research 19(9): 1655-1664.
- Talevich E, Shain AH, Botton T, Bastian BC (2016) CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLOS Computational Biology 12(4): e1004873.
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics 38(8): 904-909.
- Pickrell JK, Pritchard JK (2012) Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genetics 8(11): e1002967.
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L (2009) Reconstructing Indian population history. Nature 461(7263): 489-494.
- Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, et al. (2014) Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505(7481): 87-91.
- Busing FMTA, Meijer E, Van Der Leeden R (1999) Delete- m Jackknife for Unequal m. Statistics and Computing 9: 3-8.
- Bai H, Guo X, Narisu N, Lan T, Wu Q, et al. (2018) Whole-genome sequencing of 175 Mongolians uncovers population-specific genetic architecture and gene flow throughout North and East Asia. Nature Genetics 50(12): 1696-1704.
- Browning BL, Browning SR (2013) Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194(2): 459-471.
- Browning BL, Browning SR (2011) A fast, powerful method for detecting identity by descent. American Journal of Human Genetics 88(2): 173-182.
- Xu S, Huang W, Qian J, Jin L (2008) Analysis of genomic admixture in Uyghur and its implication in mapping strategy. American Journal of Human Genetics 82(4): 883-894.
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, et al. (2011) Mapping copy number variation by population-scale genome sequencing. Nature 470(7332): 59-65.
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, et al. (2010) Origins and functional impact of copy number variation in the human genome. Nature 464: 704-712.
- Rodriguez-Flores JL, Fakhro K, Agosto-Perez F, Ramstetter MD, Arbiza, L, et al. (2016) Indigenous Arabs are descendants of the earliest split from ancient Eurasian populations. Genome Research 26(2): 151-162.
- Raveane A, Aneli S, Montinaro F, Athanasiadis G, Barlera S, et al. (2019) Population structure of modern-day Italians reveals patterns of ancient and archaic ancestries in Southern Europe. Science Advances 5(9): eaaw3492.
- Hunter-Zinck H, Musharoff S, Salit J, Al-Ali KA, Chouchane L, et al. (2010) Population Genetic Structure of the People of Qatar. American Journal of Human Genetics 87(1): 17-25.
- Regueiro M, Garcia-Bertrand R, Fadhlaoui-Zid K, Álvarez J, Herrera RJ (2015) From Arabia to Iberia: A Y chromosome perspective. Gene 564(2): 141-152.
- Dumas L, Kim YH, Karimpour-Fard A, Cox M, Hopkins J, et al. (2007) Gene copy number variation spanning 60 million years of human and primate evolution. Genome Research 17(9): 1266-1277.
- Jones RJ, Tay GK, Mawart A, Alsafar H (2017) Y-Chromosome haplotypes reveal relationships between populations of the Arabian Peninsula, North Africa and South Asia. Annals of Human Biology 44(8): 738-746.
- Lupski JR (2007) Genomic rearrangements and sporadic disease. Nature Genetics 39(7 Suppl 1): S43-S47.
- Lebling RW (2009) "The Saracens of St. Tropez". Saudi Aramco World.
- Metspalu M, Kivisild T, Metspalu E, Parik J, Hudjashov G, et al. (2004) Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genetics 5: 26.
- Gonzalez AM, Garcia O, Larruga JM, Cabrera VM (2006) The mitochondrial lineage U8a reveals a Paleolithic settlement in the Basque country. BMC Genomics 7: 124.
- Redman CL (1978) The rise of civilization. From early farmers to urban society in the ancient Near East. WH Freeman and Company, San Francisco, USA, pp. 183.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.