Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
An mRNA-Binding Protein of The Ancient Protist Trichomonas Vaginalis
*Corresponding author: JF Alderete, School of Molecular Biosciences, College of Veterinary Medicine, Washington State University, Pullman, Washington, USA.
Received: June 11, 2021; Published: July 06, 2021
DOI: 10.34297/AJBSR.2021.13.001877
Abstract
An in vivo UV-crosslinking assay using lysates of Trichomonas vaginalis was used to identify a 21.5-kDa protein called RNA-binding protein 1 (RBP1) that interacts with mRNA. A single transcript was detected using Northern blots that showed RBP1 mRNA was expressed in parasites. Nitrocellulose with RBP1 blotted after electrophoresis of total proteins from high versus low-iron parasites and probed with MAb CF130C3 to RBP1 were identical. A cDNA expression library screened with CF130C3 yielded a cDNA encoding for RBP1 of 189 amino acids with Mr 21.5-kDa and pI 11.01. The nucleotide sequence of the 3'-end revealed the presence of the ATTTA (AUUUA for mRNA) putative transcript destabilizing element and poly(A)-tail. Importantly, the amino acid sequence of RBP1 disclosed the presence of ribonucleoprotein (RNP) elements found within the functional domains of known RNP RNA-binding proteins. Furthermore, RBP1 was found to have two distinct RNP RNA-binding domains (RNP-1 and RNP-2) found among eukaryotic RNA-binding proteins. Southern analysis indicated a single copy gene. In vitro RNA-binding assays using the mRNA subclones without 5'- and 3'-untranslated regions (UTRs) of the ap51-1 adhesin of T. vaginalis showed the binding of RBP1 to the 3'-UTR. Finally, in addition to T. vaginalis, RBP1 was detected among three different members of the family Trichomonidae. This is the first demonstration of a protein that binds to the 3'-UTR of transcripts identified for this ancient protist.
Keywords: Mrna-Binding Protein; Trichomonas Vaginalis; Low-Iron Parasites; Amino Acids; Ribonucleoprotein; Untranslated Regions.
Abbreviations: RBP1: mRNA-binding protein 1; MAb: monoclonal antibody; AP: plasmids poly tail; RNP: Ribonucleoprotein Elements; STIs: Sexually Transmitted Infections; UTR: Untranslated Region
Introduction
Trichomonas vaginalis is responsible for the most common, nonviral sexually transmitted infection (STI) worldwide [1]. A hallmark of this STI is the asymptomatic nature of infection among both women and men. In addition, there is an association of infection in women with the development of other genitourinary complications, including adverse pregnancy outcome [2], increased susceptibility to HIV infection [3], and cervical neoplasia [4].
Environmental regulation of numerous properties by iron is a hallmark of this ancient protist [5-9]. For example, the adhesin proteins AP65, AP51 and AP33 required for cytoadherence of trichomonads to vaginal epithelial cells are up-regulated by iron, and all adhesins are members of multi-gene families, [5-8]. Interestingly, the transcripts of the adhesin proteins differ in the 3'-untranslated regions (UTR) [5,9]. Further, several adhesin mRNA transcripts possess AU-rich elements (AREs) [5,9] known to regulate RNA instability in other systems [10]. One of the adhesins, AP51-2, was found not to respond to iron in up-regulation of expression [5]. Therefore, members within these multigene families may be differentially regulated. To date, however, no regulatory proteins have been identified that may play a role in any of these parasite responses to the extracellular environment.
With the goal of understanding the mechanism(s) of regulation of gene expression in T. vaginalis and given the knowledge base available on post-transcriptional regulation in other systems [11-13], it was important to determine if these organisms possess mRNA-binding proteins. To this end, the gene rbp1 encoding a 21.5-kDa RNA-binding protein (RBP1) was isolated, and RBP1 was shown to bind mRNA in vivo using UV-crosslinking experiments. RBP1 was present in equal amounts regardless of the iron status of parasites and was constitutively expressed. The sequence of recombinant cDNA clones indicated that RBP1 contains both primary domains and secondary structural elements characteristic of members of the RNA-binding protein family [14-16]. We show that RBP1 binds to motifs at the 3'-untranslated region (3'-UTR) of the mRNA of adhesin protein AP51-1. Thus, this work established an initial framework into the regulation of T. vaginalis gene expression.
Methods
Culture and growth of microorganisms and electrophoresis
The fresh clinical Trichomonas. vaginalis isolate T016 was grown in normal, high-iron, or low-iron Trypticase-yeast extract-maltose (TYM) medium with 10% heat-inactivated horse serum, as before [5,7,8]. Other species of trichomonads were grown similarly. Recombinant Escherichia coli InvαF' with plasmids pCDNAII or pCR2.1 (Invitrogen Corp., Carlsbad, CA) were grown in Luria Broth (LB) or on LB agar plates with 65 μg/ml ampicillin [17,18]. The vector pCDNAII was used to generate the cDNA library [17], and all PCR products were cloned into pCR2.1. Induction of RBP1 expression in recombinant E. coli was done with 1 mM isopropyl-β-D-thiogalactopyranoside (Sigma Chemical Co., St. Louis, MO, USA).
In vivo UV-irradiation and purification of the mRNA complex
Parasites (3 × 108) washed three times with cold PBS were suspended in 1 ml PBS containing 100 μM Nα-p-tosyl-L-lysine chloromethyl ketone (TLCK) (Sigma). Organisms were placed in individual wells of 24-well culture plates and then exposed for 5 minutes (min) at 4°C to a wavelength of 254 nm at 4.5cm. The radiation dose was ~6.5 × 103 ergs/mm2. Organisms were centrifuged and suspended in 9 ml lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 1.5 mM MgCl2, 1% SDS, and 1% β-mercaptoethanol) in the presence of 5 U/ml RNase Block (Stratagene USA, La Jolla, CA) [17]. TLCK (1 mM) and Leupeptin (100 μg/ml) (Sigma) were added followed by addition of Triton X-100 (0.5% final concentration). Complete lysis of trichomonads was achieved by passaging three times through a sterile 18-gauge needle and examining by darkfield microscopy. The lysate was incubated at 90°C for 5 min followed by ice for 2 min, and then 0.5 M LiCl and 70 mg oligo(dT)-cellulose (Thermo Fisher Scientific, Waltham, MA USA) were added. The mixture was incubated overnight at 4°C with constant agitation followed by washing the oligo(dT)-cellulose with 70 ml of 10 mM Tris-HCl, pH 7.4, containing 500 mM LiCl prior to transfer to an RNA spin column (Thermo Fisher) for elution of mRNA-protein complexes with 10 mM Tris-HCl, pH 7.4. RNA was digested with 10 U RNase cocktail (Stratagene, La Jolla, CA USA) for 1 hour (h) at 37°C. Proteins were precipitated overnight with 10% trichloroacetic acid. The proteins were pelleted by centrifugation and washed three times with cold PBS. The final pellet was solubilized in 20 µl electrophoresis dissolving buffer [5,7,18]. After boiling for 3 min, solubilized proteins were electrophoresed as described below.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting
Proteins were electrophoresed using 10% acrylamide gels [9], and gels were stained using the ProteoSilverTM Silver Stain Kit (Sigma). For some experiments duplicate gels were electroblotted onto nitrocellulose after SDS-PAGE for immunoblotting and probing with monoclonal antibody (MAb) CF130C3 reactive with RBP1, as before [5,7,8,18].
Isolation of C23-1 cDNA encoding recombinant RBP1 and sequencing rbp1
The MAb CF130C3 to RBP1 was generated as has been extensively described [18,19]. The cDNA expression library screened using MAb CF130C3 isolated a recombinant E. coli with a cDNA insert (Figure 1 Part A), and colonies of E. coli colonies were subjected to insert analysis [5,17]. The recombinant cDNA E. coli clone labeled C23-1 encoded RBP1.
The nucleotide sequence of the 605-bp C23-1 cDNA (Figure 1) was obtained using the T7 Sequenase® Version 2.0 7-deaza-dGTP DNA sequencing kit (Amersham Life Science, Arlington Heights, IL). The C23-1 cDNA was missing 5'-end information, which was obtained using the MarathonTM cDNA Amplification kit (Clontech Laboratories, Inc., Palo Alto, CA) with the antisense primer P1 (5'-GGTTCTTGATAGCGGCCATAAGGGC-3') (Figure 1, dotted underline). This additional nucleotide sequence still lacked the 5'-end transcriptional and translational start sites, which was then derived using the Universal GenomeWalkerTM (Clontech) (Figure 1, italics and underlined) using the P1 primer and antisense primer P2 (5'-CGAAACCACGGGAAATGATCTGGCC-3') as per the manufacturer’s instructions for mRNA and genomic DNA, respectively. Computer analysis of acquired sequences was carried out using PC/GENE® (Release 18.0) (IntelliGenetics, Inc., Mountain View, CA) and Omiga 1.1 (Oxford Molecular Group Ltd., Campbell, CA). Blast homology searches utilized SwissProt, GenBank, and EMBL.
Northern and Southern hybridization
Southern analysis was performed, as before [5,17,18,20], on purified genomic DNA digested to completion with AvaI, EcoRI, and EcoRV (Figure 2) (New England Biolabs, Inc., Beverly, MA) that do not cut within the DNA. Restricted DNA (10μg) was electrophoresed and transferred to Zeta-probe membrane (Bio-Rad Laboratories, Richmond, CA, USA) as recommended by the manufacturer. Hybridizations were performed with nick-translated 32P-labeled C23-1 cDNA. Blots were prehybridized using 50% formamide, 120 mM Na2HPO4, 250 mM NaCl, 7% SDS and 1 mM EDTA) at 42°C for 18h. Sequential washing of membranes was then performed with 2X SSC (1X SSC is 250 mM NaCl plus 15mM Na3-citrate, pH7.0) with 0.1% SDS, 0.5X SSC-0.1% SDS, and 0.1% SSC-0.1% SDS. Incubations were at room temperature (RT) for 15, 30 and 30 min, respectively.
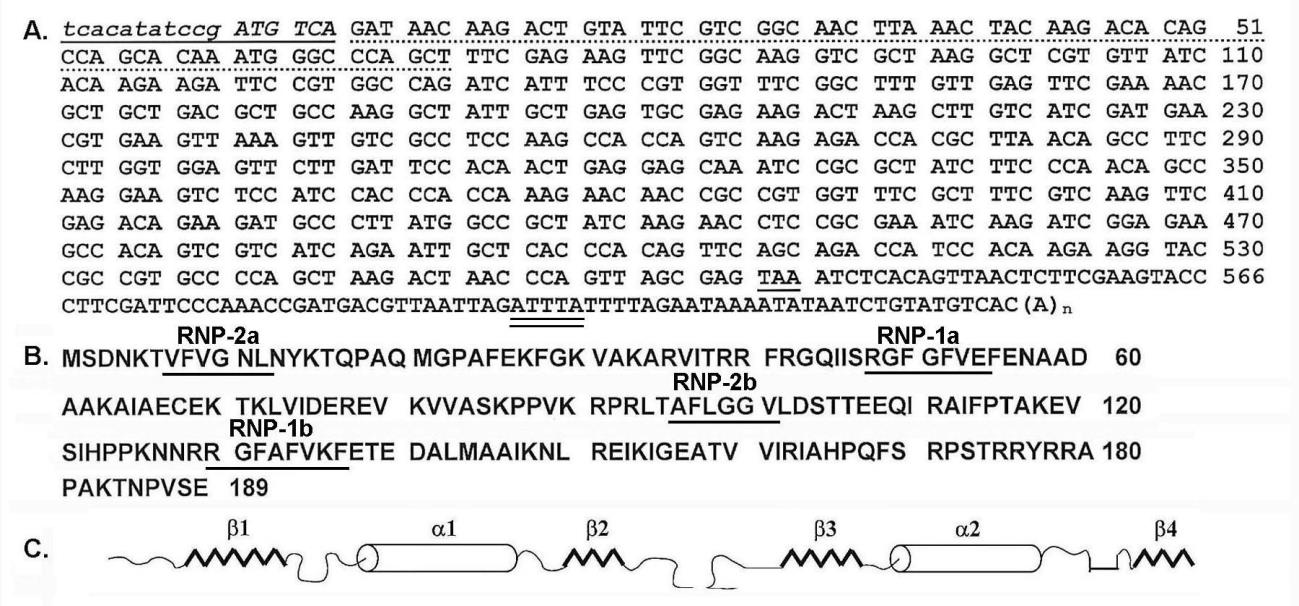
Figure 1: Complete nucleotide and amino acid sequences of RBP1. Part A is the 605-bp nucleotide sequence of C23-1 cDNA within which is the 566-coding sequence of RBP1 from start codon (ATG) to stop codon (TAA). The underlined and italics in addition to the dashed sequence were missing in the original cDNA clone and were acquired as described in Methods. The 3’-UTR nucleotide sequence shows a putative AU-rich element (ATTTA) (double underlined, AUUUA for mRNA) and the corresponding poly(A) tail. Part B is the 189 amino acid sequence with the corresponding ribonucleotide-binding sequences labeled RNP 1a, RNP 1b, RNP-2a, and RNP-2b. Part C is the predicted secondary structure with the βαββαβ organization consistent with that reported for other RNA-binding proteins [14].
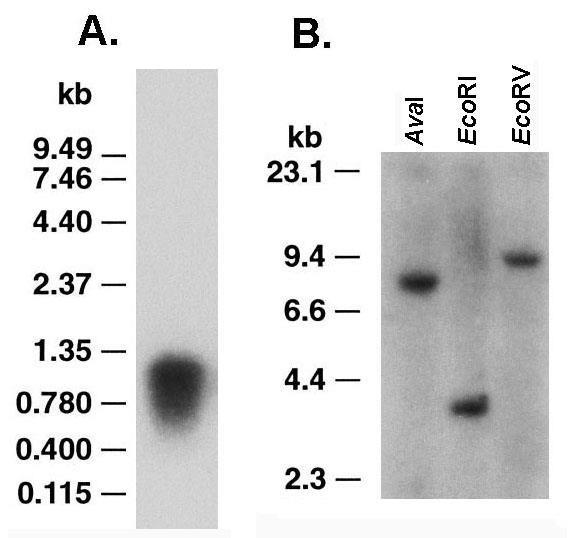
Figure 2: Northern and Southern analyses of RBP1. Part A Northern hybridization using 10 µg of RNA electrophoresed in 1% denaturing agarose get followed by blotting onto Zeta-probe membrane. Hybridization was under high stringency conditions with nick-translated, 32P-labeled C23-1 cDNA. The single band of ~1-kbp in size encodes for RBP1. Part B is of Southern analysis of genomic DNA digested to completion with restriction enzymes, as indicated, and electrophoresed in 0.8% agarose gel followed by transferring to Zeta-probe membrane. Hybridization under high stringency with 32P-labeled C23-1 cDNA revealed single bands indicative of the single copy nature of rbp1.
Methods for Northern analysis was performed as before [5,17,20]. Purified RNA obtained by FastTrack® 2.0 as specified by the manufacturer (Thermo Fisher) was from trichomonads grown in normal TYM-serum medium. RNA (10μg) was electrophoresed in 1% denaturing agarose and then transferred onto Zeta probe membrane for hybridizing at high stringency conditions with nick-translated 32P-labeled C23-1 cDNA.
Determination of mRNA-binding location by RBP1 using competitors in UV-crosslinking assays using RBP1 in lysates and recombinant RBP1 in recombinant E. coli extracts
To show the location of RBP1 binding to mRNA in competition experiments, the T. vaginalis adhesin protein ap51-1 gene [5] was used to generate the full-length ap51-1 transcript and ap51-1 subclones (Table 1). Poly(A), poly(C), and poly(U) RNA homopolymers (Sigma) were included in separate competition experiments. Linearized plasmids (1 μg) from the ap51-1 subclones were then in vitro-transcribed. To generate the full-length, radiolabeled SC5.3 RNA to use as probe in the competition experiments (Table 1), 50 μCi 32P-UTP (NEN) was included in the reaction. RNAs were ultimately passed through MicroSpinTM G-25 columns (Amersham Pharmacia Biotech, Inc., Piscataway, NJ).
All in vitro RNA competition experiments used either lysates of parasites or French press extracts of E. coli expressing the cDNA C23-1 that encodes for recombinant RBP1 [3]. Competitor, unlabeled RNA (2 pmol) was added to either lysate or extract and incubated for 10 min at RT followed by addition of 20 fmol radiolabeled RNA probe in 10 mM Tris-HCl (pH 8.0), 15 mM KCl, 1 mM MgCl2, 5 U RNase Block, 0.2 μM DTT, 5 % glycerol, 0.2 μg tRNA, and 100 μg/ml bovine serum albumin [20]. The reactions were incubated for 20 min, and RNA was crosslinked to RBP1 by exposure to UV for 5 min on ice. The mixture was then digested with 10 U RNase Cocktail for 1h at 37°C. The proteins were subjected to SDS-PAGE for autoradiography. The intensity of bands in X-ray films were scanned for quantitation.
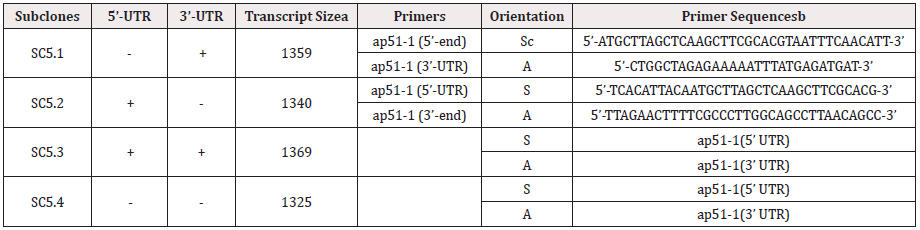
Table 1: Summary of ap51-1 adhesin subclones, orientation and primers.
Abbreviations: UTR: Untranslated Region; S: Sense Strand; A: Antisense Strand; ATG; Start Codon
a. Transcript size is given as number of bases.
b. Primers corresponding to the 5’-end of each gene (S) include ATG start codon (bold). Primers for the 5’-ends of clones SC5.2 and SC5.3
also contain the Inr-like promoter element (underlined) [27].
c. Sense (S) and antisense (A) primers are relative to the ap51-1 gene as described before [5].
PCR of trichomonads
Genomic DNA from T. vaginalis, T. suis, T. foetus KV1, T. tenax, and Pentatrichomonas hominis was isolated as described above and before [20]. Primers derived from the rbp1 gene included the P1 primer and the sense primer P3 (5'-CCTCCAAGCCACCAGTCAAGAGACC-3'). The hot start protocol (94°C for 1 min) for PCR used the TaqStart Antibody (CLONTECH). Products were amplified during 30 cycles of PCR at 94°C for 30 sec, 60°C for 30 sec, and 68°C for 2 min, and cloned directly into pCR2.1 for sequencing (Thermo Fischer).
Nucleotide sequence accession number
The GenBank accession number for the nucleotide sequence of rbp1 obtained in this study is AF084572. The accession number for the ap51-1 adhesin gene of T. vaginalis is U87093.
Results
Identification of a T. vaginalis RNA-binding protein
We identified the RNA-binding protein RBP1 from T. vaginalis by performing in vivo UV crosslinking experiments using organisms grown in normal TYM-serum medium. As can be seen in Figure 3, the 21.5-kDa protein band labeled RBP1 was detected in the fraction eluted from the oligo(dT)-cellulose column in the UV-treated samples (lanes 1). No protein was visible in non-UV-irradiated organisms (lane 2). All other bands corresponded to the RNase cocktail used to treat the samples.
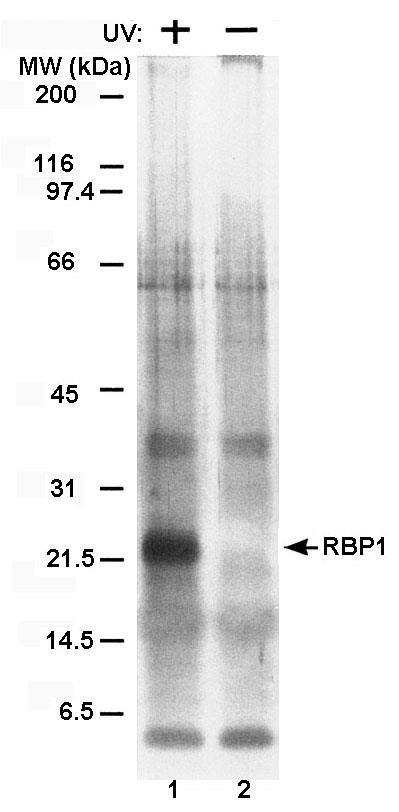
Figure 3: RBP1, an RNA-binding protein from T. vaginalis, is readily detected utilizing the in vivo UVcrosslinking assay. Silver-stained gels after SDS-PAGE of proteins purified by oligo(dT)-cellulose chromatography of lysates from parasites treated (+) and untreated (-) with UV irradiation prior to solubilization. Lysates were derived from parasites grown in normal TYM-serum medium (Methods). Lanes 1 and 2 show the presence and absence of the protein band representing RBP1 from the UV-treated and untreated samples, respectively. Numbers on the left refer to the molecular weight (MW) in kilodaltons (kDa) of protein standards.
MAb CF130C3 reacts with RBP1
Figure 4 (part A) repeats a UV crosslinking experiment and shows RBP1 in the silver-stained band after SDS-PAGE (lane 1). A duplicate gel as lane 1 was immunoblotted onto nitrocellulose, and MAb C130C3 readily detected the same intensity of the protein as can be seen in lane 3. No corresponding protein was detected in the absence of UV treatment for both stained gels and nitrocellulose blots (lanes 2 and 4). Part B presents an immunoblot of total proteins of T. vaginalis grown in either high- (lane 1) and low-iron (lane 2) medium, and identical intensities of RBP1 protein with MAb CF130C1 were obtained, suggesting constitutive expression of RBP1 regardless of iron status of organisms. Finally, an immunoblot was performed comparing total proteins of T. vaginalis with recombinant E. coli extract from clone labeled C23-1 that was reactive with MAb CF130CF. As can be seen in part C a band of 21.5-kDa of the natural T. vaginalis protein (lane 1) and a recombinant E. coli protein of ~25-kDa for the cDNA-encoded protein (lane 2) were detected. The slightly higher MW of the C23-1 protein resulted from fusion of RBP1 with β-galactosidase. As a control no protein was reactive with MAb in E. coli with vector alone (not shown).
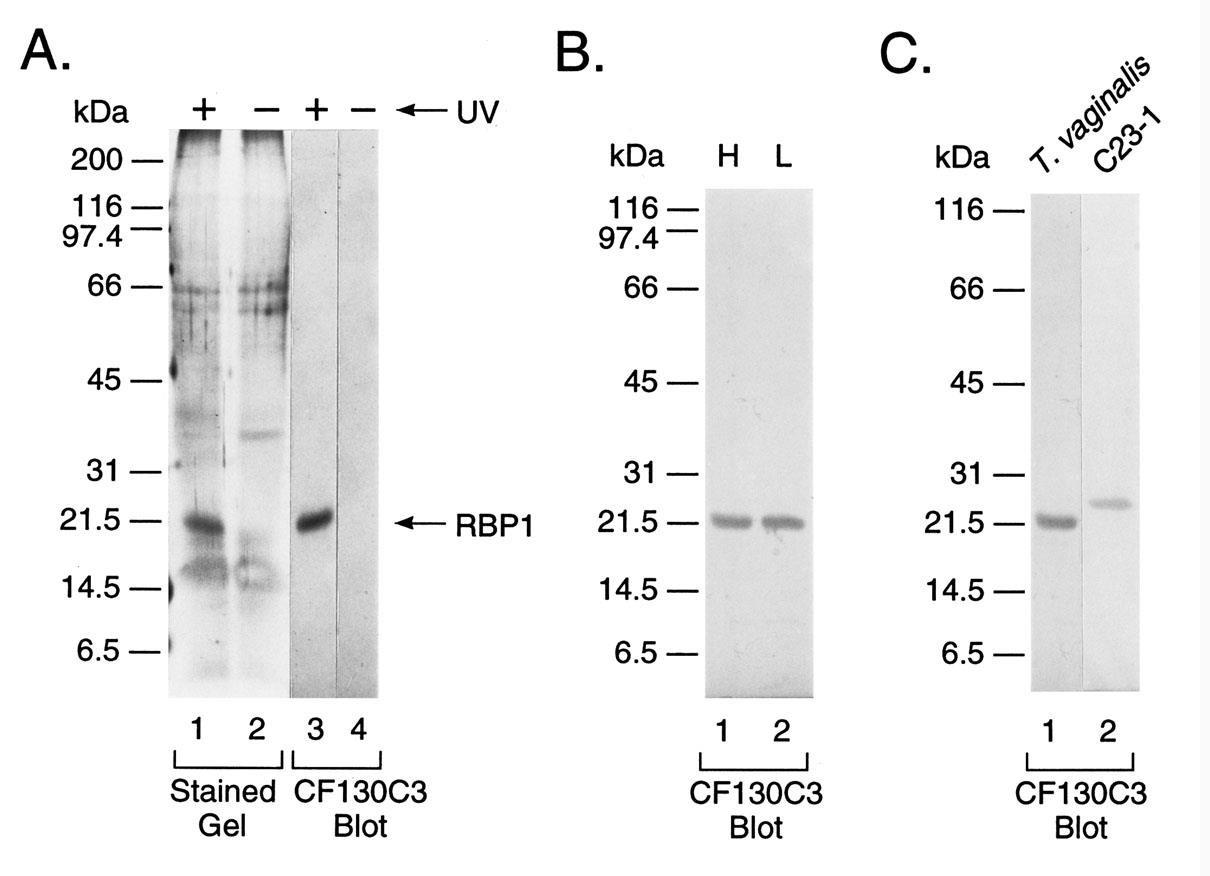
Figure 4: Immunoblot detection of RBP1 with mAb CF130C3. Part A presents the presence (+) or absence (-) of UV irradiation of lysate of T. vaginalis grown in normal TYM-serum medium (Methods) followed by SDSPAGE and silver staining of gels (lanes 1 and 2) and the reaction of duplicate gels immunoblotted onto nitrocellulose and probed with MAb CF130C3 (lanes 3 and 4). Only irradiated lysates yielded a silver-stained protein band (lane 1), which was detected by MAb CF130C3 (lane 3). Importantly, the additional protein bands seen in silver-stained gels are from the RNase cocktail used to treat the samples. Part B presents MAb CF130C3 binding to RBP1 blotted onto nitrocellulose after SDS-PAGE of UV-irradiated lysates for T. vaginalis grown in iron-replete (lane 1) versus iron-depleted medium (lane 2). Identical intensities of bands were evident regardless of the iron status of parasites. In part C the mAb CF130C3 is used as probe to detect RBP1 after SDS-PAGE and immunoblotting onto nitrocellulose from T. vaginalis lysate (lane 1) and recombinant E. coli extract (Methods) (lane 2). The higher Mr of the recombinant protein is due to the fusion of C23-1 cDNA to β-galactosidase.
Nucleotide, amino acid sequence of RBP1 and similarity to other ribonucleoproteins (RNPs) that bind RNA
Figure 1 (part A) presents the nucleotide sequence of the 605-bp C23-1 cDNA, which includes the 566 nucleotides coding for RBP1 from the ATG start codon to the TAA stop codon. Initial sequencing revealed an incomplete 5'-end of the gene and the Inr element [21]. The additional 66 bases (dash underlined) and the 17 bases of the Inr element and start codon and an additional codon (underlined italics) were obtained as described in the Methods section. Interestingly, the 3'-UTR nucleotide sequence had a putative AU-rich element (ARE) (ATTTA) (double underlined, AUUUA for mRNA) reported to confer instability to mRNA species in other eukaryotes [10] and had a corresponding poly(A) tail.
Part B shows the protein consisted of 189 amino acids with an Mr of 21,143.51 and a theoretical pI of 10.30. Database analysis revealed RBP1 to possess two eight amino acid ribonucleotide binding sequences labeled RNP-1a (amino acids 48-RGFGFVE-54) and RNP-1b (129-RGFAFVKF-136), which have consensus sequences to other RNA-binding domains [14]. Two other sequences designated RNP-2a (7-VFVGNL-12) and RNP-2b (96-TAFLGGV-101) were also identified. There are two ribonucleotide-binding domains (RBDs) with RBD1 from amino acid 1 to 85 and RBD2 from 90 to 167. The sequences of the RBD1 and RBD2 do not possess homology at the amino acid level either to each other or to other RNA-binding proteins [14]. Finally, the amino acid sequence indicates a lack of a transmembrane domain.
Furthermore, Part C illustrates the secondary structure predicted for each RBD and consists of a βαββαβ organization, as has been reported for other RNA-binding proteins [14]. These data indicate that upon protein folding, the structure is expected to have a four-stranded anti-parallel β sheet on which lie two perpendicular α-helices [16], an arrangement that may occur twice in the RBP1 relative to the four RNP sequences.
Hydrophilicity-hydrophobicity profile of RBP1 and alignment with ribonucleotide RNA-binding domains (RBD)
Figure 5 (part A) shows the hydrophobicity-hydrophilicity profile of RBP1. This plot of RBP1 was obtained using the Expasy ProtScale (https://web.expasy.org/protscale/), which computes the profile produced by amino acids. This plot further illustrates that the RNA-binding sequences of the two RBDs (part B) fall exactly within hydrophilic regions of the protein (part A). Finally, part A shows the position of the two tyrosine residues and of cysteine. In preliminary experiments, tyrosine at position 177 is phosphorylated (not shown). Not unexpectedly, treatment of RBP1 with NTCB (Methods) cleaved the protein at the cysteine position 68 into two distinct fragments of 7.4-kDa and 13.8-kDa.
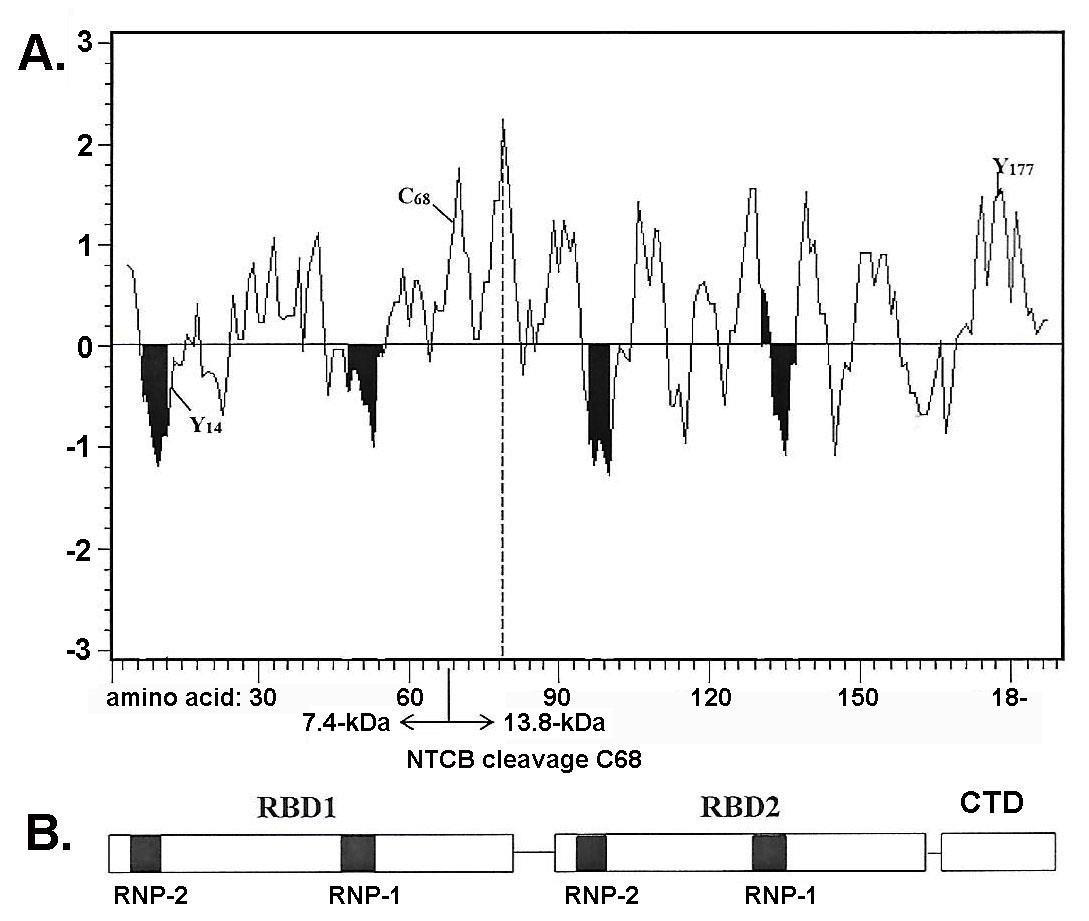
Figure 5: Hydrophilicity-hydrophobicity profile of RBP1 with ribonucleotide RNA-binding domains (RBD1 and RBD2). Part A is of the plot of RBP1 of hydrophobicity (positive values) with the shaded areas corresponding to the ribonucleotide-binding sequences labeled RNP 1a, RNP 1b, RNP- 2a, and RNP-2b shown in part B. RBP1 was treated with NTCB, which reacted with cysteine at amino acid 68 (Methods), which gave the expected 7.4-KDA and 13.8-kDa peptides after SDS-PAGE and silver staining. The tyrosine at amino acid 177 is noted because of phosphorylation.
The mRNA transcript and single copy of the rbp1 gene
We next performed Northern analysis of 10 μg purified RNA derived from trichomonads grown in normal TYM-serum medium. The RNA was electrophoresed in 1% denaturing agarose followed by blotting onto Zeta probe membrane and probing with nick-translated 32P-labeled C23-1 cDNA. Figure 2 (part A) shows the radioactive probe detected a single band of ~1-kb in size and sufficient to encode the transcript for RBP1.
Next, purified genomic DNA was digested to completion with AvaI, EcoRI and EcoRV followed by electrophoresis in 0.8% agarose gel. The gel was then transferred onto Zeta-probe membrane and hybridized with nick-translated 32P-labeled C23-1 cDNA. Only single bands were obtained that show RBP1 is single copy. Genomic DNA was also digested to completion with HincII, BamHI, and HindIII, which gave similar single band patterns (data not shown). These data suggest that rbp1 is a single copy gene.
RBP1 associates with the 3’-UTR
In Figure 6 data are presented from UV-crosslinking experiments using lysates of T. vaginalis organisms (part A) and extracts of recombinant E. coli expressing RPB1(part B) to examine the region of a transcript recognized by RBP1. RNA-binding assays were performed using in vitro-transcribed ap51-1 adhesin RNA and subclones [5]. Table 1 presents the cloning of ap51-1 with both 5'-UTR and 3'-UTR (SC5.3), without both 5'-UTR and 3'-UTR (SC5.4), without 5'-UTR only (SC5.1), and without 3'-UTR (SC5.2) only. The complete ap51-1 transcript (SC5.3), containing both UTRs, was generated, radiolabeled, and used as probe. The remaining ap51-1 subclones and RNA homopolymers were not radiolabeled and used as competitors in binding assays.
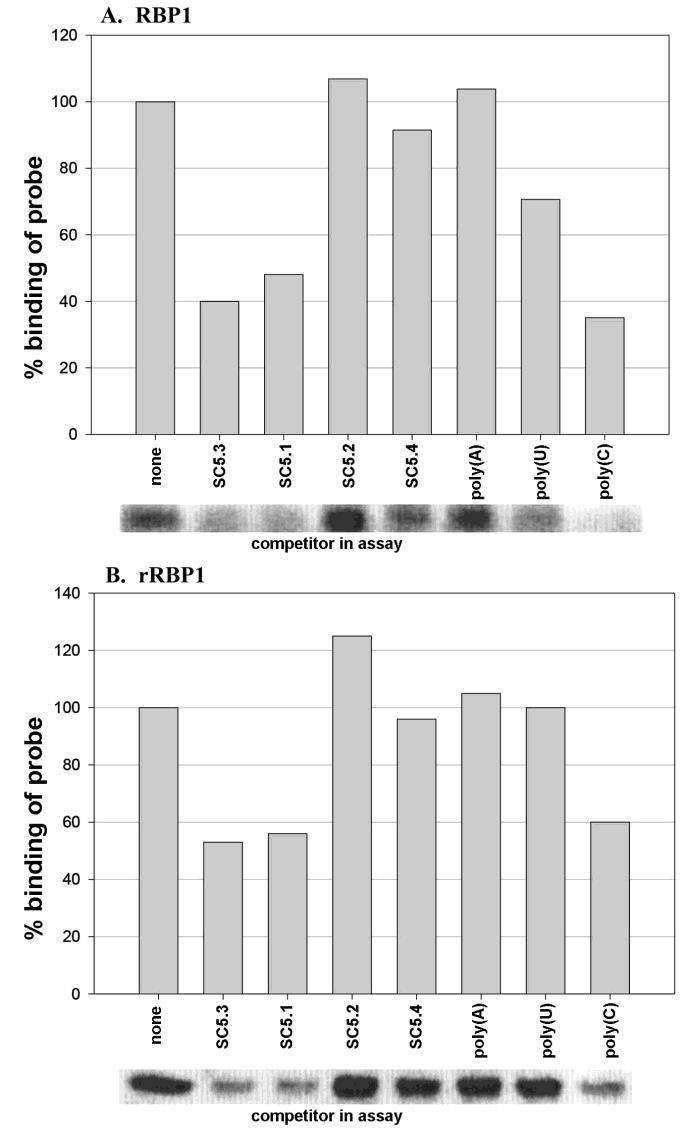
Figure 6: RBP1 interacts preferably with the 3'-UTR in UV crosslinking experiments using T. vaginalis lysates (part A) or with recombinant E. coli extracts with subclone C23-1 encoding RBP1. For these experiments RNA-binding assays were performed using in vitro-transcribed ap51-1 adhesin RNA reported before for the trichomonad AP51 adhesin protein [5]. Unlabeled AP51 subclones for competition experiments (Table 1) were of ap51-1 with both 5'-UTR and 3'-UTR (SC5.3), without both 5'-UTR and 3'-UTR (SC5.4), with 5'-UTR only (SC5.2) and with 3'-UTR only (SC5.1). Unlabeled RNA homopolymers poly(A), poly(U), and poly(C) were also used as competitors. The complete ap51-1 transcript (SC5.3), containing both UTRs, was generated, radiolabeled, and used as probe. For both parasite lysates (A) and bacterial extracts expressing RBP1 (B) only SC5.3, SC5.1 and poly(C) competed for binding with the radiolabeled ap51 transcript.
Figure 6 (part A) shows the experiment with parasite lysates. SC5.3 and SC5.1 RNAs with 3'-UTRs compete with binding of radiolabeled SC5.3 to RBP1 by 60% and 52%, respectively. SC5.2 RNA with the 5'-UTR including the Inr (Figure 1) and SC5.4 without both 5'-UTR and 3'-UTR were ineffective competitors. The data presented in part B using E. coli extract with C23-1 recombinant RBP1 (Figure 4C) shows identical results. Importantly, no proteins generated in E. coli transformed with vector alone bound the T. vaginalis RNA probe. Interestingly, poly(C) RNA homopolymer but not poly(A) or poly(U) RNA competed with SC5.3 for binding to both natural RBP1 and recombinant RBP1 by 65% and 40%, respectively. Therefore, these data indicate that RBP1 recognizes motifs found within the 3'-UTR.
rbp1 is not unique to T. vaginalis
Finally, we determined rbp1 was conserved within the family Trichomonidae. PCR was performed using genomic DNA from Pentatrichomonas hominis (intestinal), Tritrichomonas foetus (bovine), T. tenax (oral) and T. vaginalis. Primers P1 and P3 were as described in Methods. As can be seen in Figure 7, PCR products of identical size were amplified from all four genomes. Each of these products was subsequently cloned and sequenced. The sequence data showed the PCR products were identical to the sequence obtained for T. vaginalis (Figure 1 Part A).
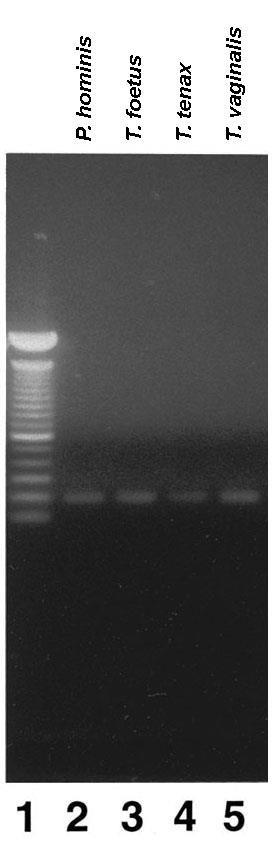
Figure 7: rbp1 is present in other Trichomonidae family members. Genomic DNA from the different trichomonads was subjected to PCR using the C23-1-derived primers P1 and P3 (Methods). PCR products were electrophoresed in 1% agarose and stained with ethydium bromide. Reactions were performed with DNA from T. suis, T. foetus, P. hominis, and T. vaginalis. Lane 1 represents to 100-bp DNA ladder. The DNA sequence of the PCR product for all species were identical (not shown).
Discussion
In this report the T. vaginalis RBP1 was the only protein detected by the in vivo UV-crosslinking assay from T. vaginalis regardless of the iron status or parasites (Figure 4), indicating that RBP1 is constitutively expressed. This single copy gene is 566 nucleotides and encodes for a protein of 189 amino acids in length. Consistent with other known ribonucleotide-binding proteins (RNPs), RBP1 also had consensus sequences to other RNA-binding domains (RBDs) [14], and the two RNA-binding domains (RBD1 and RBD2) had the secondary structure predictions of the βαββαβ organization [14,16]. Although extremely diverse in function, RNPs have conservation of primary, secondary, and tertiary structure within each RNP domain [14]. Each of the two RBDs of 1 to 85 and 90 to 167 amino acid sequences contains two RNP elements and conserved hydrophobic residues required for proper folding of the peptide and RNA-binding activity [14,15]. Although requiring experimental verification, the initial characterization of RBP1 appears to have the features predicted for these proteins. The sequence data may indicate that this RNA-binding protein also has a four-stranded anti-parallel β sheet on which lie two perpendicular α-helices (Figure 1, part C) as reported for other RBPs [14,15].
The data using full-length subclones of the T. vaginalis adhesin protein AP51 (Table 1 and Figure 6) indicates that RBP1 interacts with the 3'-UTR of mRNA transcripts. It is noteworthy that the transcripts of the adhesin proteins of T. vaginalis differ in the 3'-untranslated regions (3'-UTR) [5,9]. Thus, it will be important to examine the interaction of RBP1 with the distinct transcripts to delineate at the molecular level the exact sequences recognized by this RNA-binding protein. Interestingly, the 3'-UTR nucleotide sequence had a putative AU-rich element (ARE) (ATTTA) reported to confer instability to mRNA species in other eukaryotes [10] and had the corresponding poly(A) tail. AREs have repeatedly been shown to be bound by RBPs, resulting in altered transcript stability [10,22]. Located within 3'-UTRs, this motif has also been found in several T. vaginalis transcripts [5,17,20], although its function remains unknown. For example, it has been reported that transcripts for at least three adhesins and the phenotypically varying immunogen P270 possess either the AUUUA pentamer or UUAUUUAUU nonamer ARE in their 3'-UTRs [3,9,20]. This may suggest that the rbp1 3'-UTR with this motif may have functional significance (Figure 1).
It is known that one of the four RNA-binding domains of yeast RBP associates with poly(A) sequences in vivo [23] and interacts with eukaryotic translation initiation factor 4G (eIF4G) [24]. That RBP1 interacts with poly (A) cannot be excluded, and future work is needed to further define the exact binding sites by which RBP1 associates with mRNAs of T. vaginalis. Although speculative, the basic nature and high pI of 11.01 of RBP1 may promote associations of this protein with RNA. In addition, many characterized RBPs interact with other proteins [24-27]. Therefore, identification of proteins binding to RBP1in trichomonads is needed to further understand the function of RBP1 in vivo. It should be noted that proteins unrelated to RBP1 that interact with RNA stem loop structures and that may be involved in regulatory functions have been identified in T. vaginalis [28,29].
Finally, that RBP1 has been identified in members of the family Trichomonidae, all primitive amitochondriate protists, may suggest that this is a product early in evolution. Further, this identical conserved rbp1 gene among the Trichomonidae (Figure 7) may indicate that this gene was acquired in an ancestor common to all members of the family.
Conclusion
This report shows the identity of RBP1, a 21.5-kDa RNA-binding protein that interacts with the 3'-UTR of the ap51-1 adhesin of T. vaginalis. Synthesis of RBP1 was unaffected by the levels of iron during growth and is constitutively expressed. Two distinct ribonucleotide-binding domains (RBDs) were identified in RBP1. RBD1 from amino acid 1 to 85 and RBD2 from 90 to 167 had no homology at the amino acid sequence level within the protein and with other RNA-binding proteins [14]. The secondary structure predicted for each RBD consisted of a βαββαβ organization, as has been previously reported [14]. RBP1 is without a transmembrane domain. The single copy rbp1 gene is present among different members of the family Trichomonidae. This is the first RNA-binding protein identified for this ancient protist.
Acknowledgement
This study was supported by Public Health Service grants AI-43940 and AI-39803 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health. As per instructions by the International Committee of Medical Journal Editors on “Who Is an Author?,” (http://www.icmje.org/ recommendations/browse/roles-and-responsibilities/definingthe- role-of-authors-and-contributors.html), I want to acknowledge Erica Wargo who generated the data under my supervision in my laboratory while at The University of Texas Health Science Center at San Antonio, Texas.
Conflict of Interest
I declare that there are no conflicts of interest. I alone designed the study and was responsible in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Centers for Disease Control (2021) Prescription Drug Monitoring Programs (PDMPs).
- Peirce G, Smith M, Abate M, Halverson J (2012) Doctor and Pharmacy Shopping for Controlled Substances. Medical Care 50(6): 494-500.
- Keith DA, Shannon TA, Kulich (2018) The prescription monitoring program data: What it can tell you. The Journal of the American Dental Association (JADA) 149(4).
- Felter C (2020) The US Opioid epidemic. Council on Foreign Relations
- Wilson N, Kariisa M, Seth P, Smith H, Davis NL, et al. (2020) Drug and Opioid-Involved Overdose Deaths -United States, 2017-2018. Morbidity and Mortality Weekly Report (MMWR) 69(11).
- National Institute on Drug Abuse (NIH) (2020) Indiana Opioid-Involved Deaths and Related Harms.
- Meldrum, Marcia L (2016) The Ongoing Opioid Prescription Epidemic: Historical Context. American Journal of Public Health 106(8): 1365-1366.
- Mann B (2020) Federal Judge Approves Landmark $8.3 Billion Purdue Pharma Opioid Settlement.
- Indiana Government (2020) Indiana Drug Overdose Dashboard.
- Brady J, Wunsch H, Di Maggio C, Lang B, Giglio J, et al. (2014) Prescription Drug Monitoring and Dispensing of Prescription Opioids. Public Health Reports 129(2): 139-147.
- Pew Charitable Trusts (2016) Prescription Drug Monitoring Programs.
- Indian Health Services (2021) Opioids and the Covid-19 pandemic: Addressing the opioid crises during the Covid-19 pandemic.
- Deangelis C (2016) Big Pharma Profits and the Public Loses. The Milbank Quarterly 94(1): 30-33.
- Jones MR, Viswanath O, Peck J, Kaye AD, Gill JS, et al. (2018) A Brief History of the Opioid Epidemic and Strategies for Pain Medicine. Pain Medicine Pain Ther 7(1): 13-21.
- The United States Department of Justice (2020) Opioid Manufacturer Purdue Pharma Pleads Guilty to Fraud and Kickback Conspiracies.
- Carise D (2018) What we can do to stop patient brokering: Meaningful change can be accomplished through legislation, increased public awareness and at the treatment level. Behavioral Healthcare Executive 38(4): 15.
- Brill A, Ganz S (2018) The geographic variation in the cost of the opioid crisis. American Enterprise Institute.
- Keogh JG, Fink JL (2020) Recent law addresses the opioid crisis. Pharmacy Times 88(8).
- Rhodes E, Wilson M, Robinson A, Hayden J, Asbridge M, et al. (2019) The effectiveness of prescription drug monitoring programs at reducing opioid-related harms and consequences: A systematic review Bio Medical Central (BMC) Health Service Research 19(784).



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.