Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Identification of Novel Drug Targets and Antigenic Proteins in Helicobacter hepaticus through Proteome-Mediated Mining
*Corresponding author: Çiğdem Yılmaz, TUBITAK Gen Mühendisliği ve Biyoteknoloji Enstitüsü, Gebze, Kocaeli, 41470, Turkey
Received: April 27, 2021; Published: July 23, 2021
DOI: 10.34297/AJBSR.2021.13.001900
Abstract
Helicobacter hepaticus is a known pathobiont that causes hepatocellular carcinoma and intestinal cancer in mice. In recent studies, Helicobacter hepaticus in human patients with gallbladder cancer, biliary tract, and liver diseases has been reported. In this study, we aimed to identify therapeutic targets and vaccine candidates against the pathobiont via computational biology. The core proteome of Helicobacter hepaticus ATCC 51449 was retrieved from UNIPROT and analyzed to remove paralogs and duplicates. After identifying human non-homologous proteins and predicting subcellular localization, cytoplasmic proteins were subjected to essentiality, pathway, and druggability analysis. The analysis of essential cytoplasmic proteins resulted in six druggable targets having vital roles in peptidoglycan and lipopolysaccharide biosynthesis. For the identification of vaccine candidates, outer membrane and extracellular proteins were analyzed. After determining antigenic proteins, T and B-cell epitope prediction was carried out to uncover common epitopes for the candidates. Vaccine candidate identification revealed eleven antigenic proteins, five of which had overlapped T and B-cell epitopes that can elicit both humoral and cell-mediated immune response. Identified druggable targets and vaccine candidates might be used to develop successful treatment of infections caused by Helicobacter hepaticus.
At the start of the study, the average temperature and oxygen values were subnormal, indicating poor health status, whereas, by the end of the study, vital signs were all normal. During the study, a cohort study (proof of concept) was conducted with 14 individuals, in the TLC general population, who were identified and diagnosed as COVID-19 positive. After 6 days (on average) of “ELECTROCIDE™” administration, a shift of their COVID-19 status switched to negative, and a complete reversal of symptoms were observed several times faster than predicted by the CDC [1]. Of particular interest were oxygen levels because a previous in-vitro pilot study indicated that dissolved oxygen levels doubled with “ELECTROCIDE™” -It is postulated that this is an important mechanism promoting an immediate sense of wellness.
Keywords: Reverse vaccinology; Helicobacter hepaticus; Vaccine candidate; Drug target
Introduction
Increasing evidence has revealed the relationship between dysregulated microbiota-host interactions and many diseases such as cancer, metabolic syndrome, and inflammatory bowel diseases. Besides, specific bacterial species in microbiota can play important roles in specific clinical outcomes like gastric cancer [1]. Many studies have proven that chronic inflammation or infection can lead to neoplasms, and chronic inflammation of gastric mucosa caused by Helicobacter pylori is a well-known example that has a positive correlation with gastric carcinomas [2]. After the identification of H. pylori as the first pathogen classified as ‘group 1 carcinogen’ by the World Health Organization (WHO) in 1994, more studies have focused on Helicobacter species to identify their roles, especially in the liver and gastric diseases. In 1994, a study conducted with mice with multifocal necrotic hepatitis paved the way for the discovering a new Helicobacter spp. called H. hepaticus [3].
H. hepaticus is a natural inhabitant of mouse microbiota, and a persistent infection caused by this pathobiont leads to intestinal cancer, hepatocellular carcinoma, and chronic hepatitis in susceptible mice [4]. Although it was first considered as a mouse-specific pathogen, many studies have been reported for human cases. In a study, the pathogen was traced with nested- PCR and Western blotting in bile samples of the patients having cholecystitis, gallbladder cancer, and cholelithiasis [5]. Besides, higher antibody titers against H. hepaticus in patients with gallbladder cancer were found than the control group, and thus, its association with gallbladder cancer has been suggested [6]. Further studies have supported the pathogenicity of H. hepaticus in diseases of the human biliary tract and liver [7,8]. An in vitro study supported these findings by showing that H. hepaticus can induce acute inflammation in primary human hepatocytes and lead to hepatocellular carcinoma development [9]. Moreover, a metaanalysis revealed a higher presence of Helicobacter spp. including H. hepaticus in patients with biliary tract cancers [10]. Although a significant correlation between H. hepaticus and liver or biliary tract diseases has been not shown yet, it is a potential zoonotic pathogen and is still worthy of investigating whether chronic infection of H. hepaticus can be a risk factor for these diseases.
In the traditional approach, drug targets or vaccine candidates’ discovery for the control of pathogen-based diseases is generally based on protein identification through isolation and characterization of microorganisms. This approach has some crucial challenges, including challenging culturing conditions and a longer duration for exploration of proteins. The recent advancements in bioinformatics and computational biology opened the way for in silico identification of novel vaccine candidates and therapeutic targets faster and more cost-effectively. In reverse vaccinology, first described by Rappuoli in 2000, a pipeline has been used to reveal novel antigenic proteins by mining genomic or proteomic data [11]. Moreover, data mining can explore novel drug targets through a sequence or structural-based comparison of proteins with known targets.
In silico approach carried out by the genome or proteome of various pathogens including H. pylori were provided the identification of useful drug or vaccine targets [12]. In the current study, a proteome-mediated mining approach was used to reveal novel antigenic proteins and drug targets in H. hepaticus, which can be valuable as potential preventive and prophylactic agents to control the diseases in relation.
Methodology
Data retrieval and removal of paralogous sequences
The entire proteome of H. hepaticus ATCC 51449 was retrieved from the UNIPROT database and subjected to CD-HIT analysis to remove paralogous or duplicate proteins [13]. Sequence identity cut-off was set at 0.8 (80% identity) to exclude redundant sequences, and proteins with less than 100 amino acids were not also included. Selected non-paralogous protein sequences were further analyzed.
Selection of human non-homologs
NCBI BLASTp search was carried out for the non-paralogous proteins against non-redundant protein sequences of Homo sapiens (TaxID:9606) to identify human non-homologous proteins of H. hepaticus.
Prediction of subcellular localization
PSORTb v.3.0 (https://www.psort.org/psortb/) and CELLO v.2.5 (http://cello.life.nctu.edu.tw/) were used to predict the location of non-homologous proteins [14,15]. While outer membrane and extracellular proteins were considered for vaccine candidate analysis, cytoplasmic proteins were curated through the DrugBank database to reveal potential drug targets specific to the pathogen after essentiality analysis.
Identification of essential non-homologous proteins of H. hepaticus
Cytoplasmic proteins were considered as possible drug targets and subjected to BLASTp analysis through Database of Essential Genes (DEG) (http://www.essentialgene.org/) for the identification of indispensable proteins having roles in primary cellular functions of the pathogen [16]. The parameters, including bit score > 100 and percent identity ≥ 30%, were applied.
Analysis of metabolic pathways
Metabolic pathways of H. hepaticus and human were compared to reveal pathogen-specific pathways. Kyoto Encyclopedia of Genes and Genomes (KEGG) server was used to extract both H. hepaticus and H. sapiens metabolic pathways, which were then manually screened to select the pathways unique to H. hepaticus.
Non-homologous, essential cytoplasmic proteins of H. hepaticus were subjected to BLASTp analysis through the KEGG pathway database (https://www.genome.jp/kegg/pathway.html) and proteins listed in the pathways specific to the pathogen were considered for further analysis.
Druggability screening of essential cytoplasmic proteins
Essential, pathogen-specific cytoplasmic proteins were assessed by BLASTp against the Drugbank database using default parameters with a bit score > 100 and e-value < 0.001 to identify the novelty of proteins as targets. 3D structure modeling of the proteins was carried out in SWISS-MODEL and Phyre2.
3D structure modeling of the proteins was carried out in SWISSMODEL and Phyre2 protein fold recognition server (http://www.sbg.bio.ic.ac.uk/~phyre2) [17]. Then, they were subjected to the PockDrug analysis for pocket druggability investigation (http://pockdrug.rpbs.univ-paris-diderot.fr/) [18]. Also, protein-protein interaction analysis was performed in the STRING v11.0 database (http://string-db.org) to identify the hub proteins among putative targets according to the node degree (K ≥ 5) which represents the significant number of direct and indirect associations. Molecular weight estimation was performed by the Expasy ProtParam tool.
Vaxign analysis
Vaxign database (http://www.violinet.org/vaxign2) was used to analyze outer membrane and extracellular proteins to identify adhesin probability and Vaxign-ML score [19]. The inclusion criteria were as follows: adhesion probability >0.51, the number of transmembrane helices ≤ 1, and no similarity to mouse or human proteins. Besides, the proteins with Vaxign-ML score ≥ 90 were further selected.
Prediction of putative antigenic and virulent proteins
The Virulence Factor Database (VFDB) (http://www.mgc.ac.cn/VFs/) and VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) were used to predict virulent and antigenic properties of the selected outer membrane and extracellular proteins, respectively [20,21]. While the proteins were screened based on BLAST score ≥ 80 in VFDB, a threshold value of ≥ 0.5 was applied in the VaxiJen search. Molecular weight and theoretical pI values were estimated through the Expasy ProtParam tool.
Prediction of T-cell epitopes
The selected antigens were evaluated to find out potent T-cell epitopes via Immune Epitope Database (IEDB) server (http://tools.iedb.org/main/tcell/). Antigenic sequences indicative of MHC-I specific binding were predicted using the Stabilized Matrix Method (SMM) considering only frequently occurring alleles (http://tools.iedb.org/mhci/). The prediction output was determined according to IC50 values. Peptides with IC50 < 50 nM, < 500 nM, < 5000 nM are accepted as the indicator of high, intermediate, and low binding capacity, respectively [22]. In the study, the cut-off value was determined as IC50 < 50 nM. Parallelly, T-cell MHC-I binding epitopes were screened for their immunogenic properties (http://tools.iedb.org/immunogenicity/), and epitopes with positive immunogenicity values were picked for further analysis. MHC-II binding prediction was performed using the SMM-align (NetMHCII 1.1) method covering human HLA-DR locus (http://tools.iedb.org/mhcii/). Peptide length was defined as 15 (default value proposed by the server). Peptides were sorted by predicted IC50 value and peptides with IC50 < 50nM were figured out for the next analysis.
Prediction of B-cell epitopes
Bepipred Linear Epitope Prediction 2.0 via IEDB server was employed (http://tools.iedb.org/bcell/) to determine linear B-cell epitopes, and BepiPred-2.0 Sequential B-Cell Epitope Predictor was used for confirmation (http://www.cbs.dtu.dk/services/BepiPred/) [23]. The 0.5 default threshold was respected (Sensitivity=0.58564, Specificity=0.57158) for epitope prediction. The residues with scores above 0.5 were predicted as B-cell epitopes.
Structural modeling
The 3D structures of the antigenic candidates were modeled via SWISS-MODEL and Phyre2 web-server with default settings. Once the models were created, the Protein Preparation Wizard of Schrödinger package was employed to refine the structures. In the first step of the refınement, the models’ bond orders were assigned by using the Chemical Component Dictionary (CCD), missing hydrogens added, and het states determined by using the Epik tool of Schrödinger for pH 7 ± 2 [24]. Following, PROPKA (another package of Schrödinger) was employed for hydrogen bond assignment (at pH 7.0). In the second step of the refinement, the water molecules beyond 3 A from het groups were discarded from structures. Finally, energy minimization performed using the OPLS3e force field for each created structure [25].
Once refinement was completed, predicted epitopes were visualized on the structures via Maestro v12.4 to confirm that they are surface-exposed.
Results
Exclusion of paralogous and human homologous proteins
A large quantity of redundant sequences is present in the bacterial proteome mainly due to evolutionary duplications. Thus, a total of 1873 proteins of H. hepaticus ATCC 51449 was subjected to CD-HIT analysis to identify redundant sequences and proteins with < 100 amino acids. Two hundred fifty-five proteins were excluded after analysis, leaving 1618 proteins to be investigated. NCBI BLASTp analysis was carried out with non-paralogous proteins against human proteome to identify human homologs. Proteins having homology with the host proteins were excluded, and a final set of 1168 proteins were considered for further analysis (Table 1 in Supplemental Data).
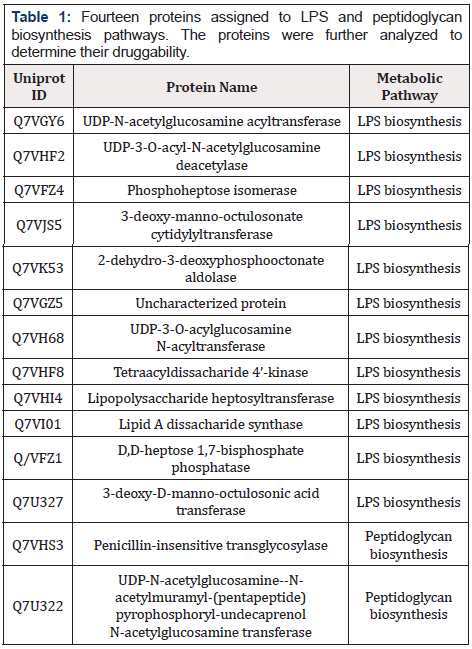
Table 1: Fourteen proteins assigned to LPS and peptidoglycan biosynthesis pathways. The proteins were further analyzed to determine their druggability.
Prediction of subcellular localization
The pool of non-homologous proteins was subjected to subcellular localization prediction using the PSORTb database. When a protein location was assigned as ‘unknown’, CELLO v.2.5 was used to overcome the limitation of PSORTb. H. hepaticus ATCC 51449 is a Gram-negative bacterium. Thus, five categories of subcellular localization were listed: (1) extracellular, (2) outer membrane, (3) periplasm, (4) inner membrane, and (5) cytoplasm. Figure 1 shows the distribution of the proteins according to their predicted subcellular locations. Among 1168 proteins, most (63%) were found in the cytoplasmic region, and the next more significant fraction (17%) localized in the inner membrane. While 9% and 4% were outer membrane and extracellular proteins, respectively, periplasmic proteins were 7% (Table 1 in Supplemental Data). Cytoplasmic, outer membrane, and extracellular proteins were subjected to further analyses since cytoplasmic proteins can serve as promising drug targets based on their pivotal roles in cell survival. In contrast, outer membrane and extracellular proteins can be considered as vaccine candidates due to their more exposed nature than cytoplasmic ones.
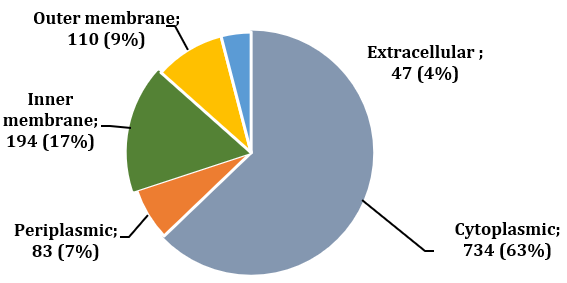
Figure 1: Distribution of the proteins based on subcellular localization predicted by PSORTb and CELLO. CELLO assessed the locations of some unknown proteins sorted by PSORTb. Cytoplasmic, outer and extracellular proteins were prioritized for the identification of drug targets and vaccine candidates.
CELLO assessed the locations of some unknown proteins sorted by PSORTb. Cytoplasmic, outer and extracellular proteins were prioritized for the identification of drug targets and vaccine candidates.
Identification of druggable cytoplasmic target proteins
The cytoplasmic proteins essentiality was analyzed in DEG 10 since they were considered as the most promising drug targets, and a total of 281 essential cytoplasmic proteins carrying out important functions in survival, adhesion, and infection were identified (Table 2 in Supplemental Data).
After retrieval of H. hepaticus and human metabolic pathways from the KEGG database, they were manually compared to reveal the pathogen-specific ones, which then resulted in 25 metabolic pathways. Analysis of 281 essential cytoplasmic proteins revealed 36 proteins having roles in these specific pathways (Table 3 in Supplemental Data) (Figure 2). Moreover, 14 proteins were assigned to peptidoglycan and lipopolysaccharides (LPS) biosynthesis pathways, which play essential roles in constructing cell walls of Gram-negative bacteria (Table 1). Therefore, to check the druggability of these fourteen proteins, they were analyzed through BLASTp search in the DrugBank database. Six of fourteen proteins matched with FDA-approved or experimental drug targets in DrugBank (Table 2) [26].
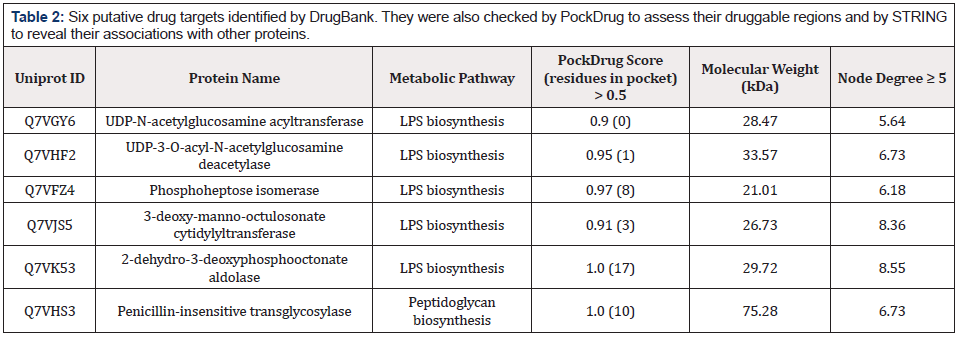
Table 2: Six putative drug targets identified by DrugBank. They were also checked by PockDrug to assess their druggable regions and by STRING to reveal their associations with other proteins.
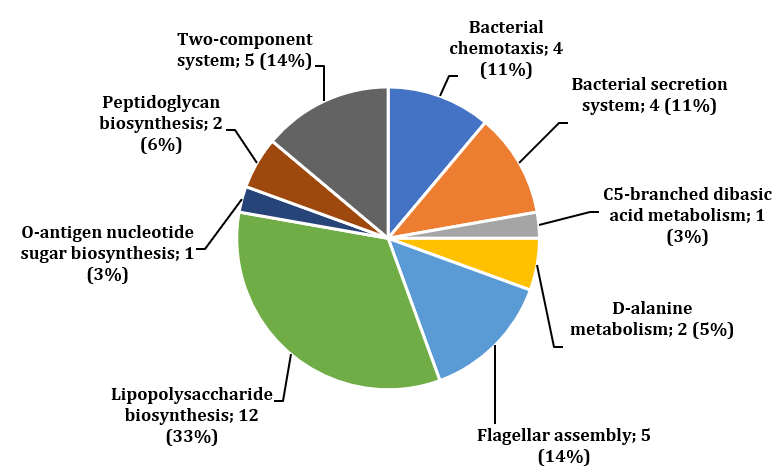
Figure 2: Distribution of essential cytoplasmic proteins based on metabolic pathways specific to H. hepaticus. The proteins included in LPS and peptidoglycan biosynthesis were selected for further analysis to identify drug targets.
After modeling and validating the protein structures, PockDrug analysis revealed that all six targets had druggable regions that a drug-like molecule can bind (Table 2). Following that, interactome analyses were performed for these six proteins to understand whether they are hub proteins representing a significant degree via interacting with many other nodes in protein networks. Interactome analyses were performed using the STRING database, and all six proteins seemed to have the node degree greater than 5 (Table 2) (Figure 3). The inhibition of hub proteins can be lethal for the survival of pathogens, so all these six proteins can be considered as novel drug targets.
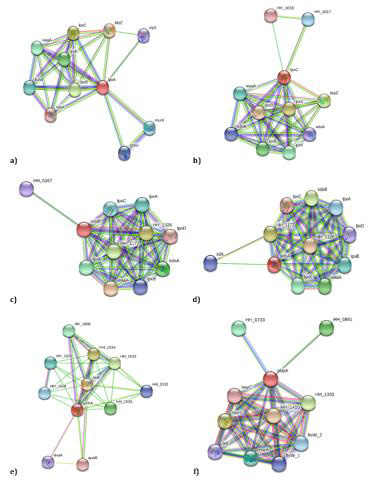
Figure 3: Protein-protein interaction diagram of the putative drug targets. Red colored circle indicates the query target. a) UDP-N-acetylglucosamine acyltransferase (lpxA), b) UDP-3-O-acyl-N-acetylglucosamine deacetylase (lpxC), c) 3-deoxy-manno-octulosonate cytidylyltransferase (kdsB), d) 2-dehydro-3-deoxyphosphooctonate aldolase (kdsA), e) Phosphoheptose isomerase (gmhA), and f) Penicillin-insensitive transglycosylase (pbpA). Among six selected targets, lpxA, lpxC, kdsA and kdsB directly interact with each other while gmhA and pbpA are in association with different nodes.
Identification of potential vaccine candidates
110 outer membrane and 47 extracellular proteins of H. hepaticus ATCC 51449 were subjected to Vaxign analysis for the identification of possible vaccine candidates having defined properties. After the analysis, the outer membrane and extracellular proteins with adhesion probability > 0.51, Vaxign-ML score ≥ 90, and the number of transmembrane helices ≤ 1 were considered as putative vaccine candidates and selected for further analysis (Table 4 in Supplemental Data)
Assessment of antigenicity and virulence of putative vaccine candidates
Potential vaccine candidates (45 outer membrane and 26 extracellular proteins) were screened for virulent and antigenic properties through Virulence Factor Database (VFDB) and VaxiJen v.2.0, respectively. 4 outer membrane and 7 extracellular proteins were listed as candidates having virulence and antigenic properties. Physiochemical properties, including molecular weight and theoretical pI values, were assessed by the Expasy ProtParam tool (Table 3).
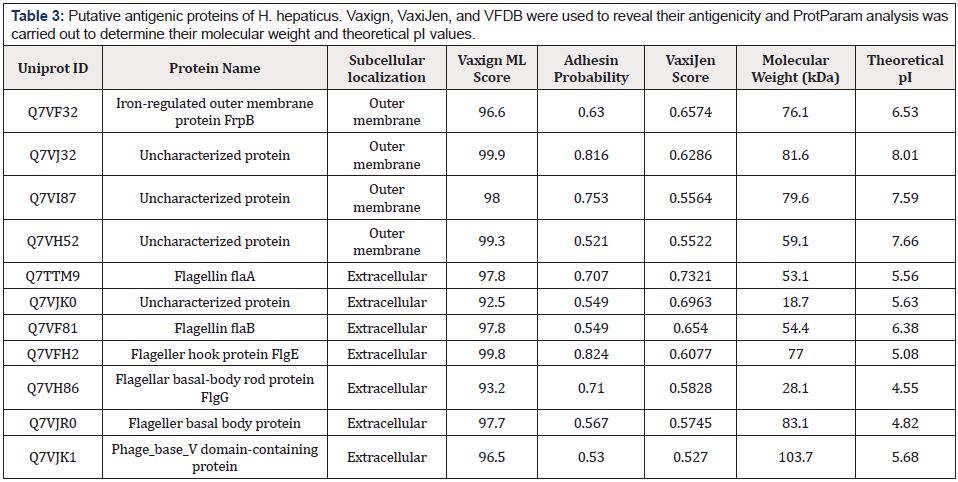
Table 3: Putative antigenic proteins of H. hepaticus. Vaxign, VaxiJen, and VFDB were used to reveal their antigenicity and ProtParam analysis was carried out to determine their molecular weight and theoretical pI values.
Prediction of potent T-cell epitopes of putative antigenic proteins
T-cell dependent cellular immune response is initiated following antigen presentation by Major Histocompatibility Complex (MHC) class I and class II. Therefore, it is inevitable to predict epitope sites of the prioritized 11 antigenic proteins for potential vaccine design. First of all, selected proteins were screened to find epitopes presenting specific binding capacity to MHC class I by IEDB server. High binding affinity property was considered equivalent to IC50 < 50 nM, and matching epitopes were furtherly subjected to immunogenicity prediction and checked for their ability to provoke an immune response. Following double screening covering MHC I-binding and immunogenicity prediction, epitopes with IC50 < 50 nM value and positive immunogenicity score were listed in Table 5 in Supplemental Data. Secondly, candidate proteins were depicted for the specific binding capacity to MHC class II by the IEDB server (IC50 < 50 nM) (Table 6 in Supplemental Data).
B-cell epitopes of putative antigenic proteins
In addition to T-cell epitope prediction, 11 proteins were examined for B-cell epitope prediction (humoral immunity). The Bepipred server was used to find linear epitopes, respecting 0.5 threshold value proposed by version 2.0. As the linear epitopes’ location is in close relation with physicochemical properties of proteins, candidate proteins were subjected to Chou and Fasman beta-turn prediction, Emini surface accessibility prediction, Karplus and Schulz flexibility prediction, Kolaskar and Tongaonkar antigenicity prediction, and Parker Hydrophilicity Prediction (Data not shown). Peptides of 5-15 amino acids in length for each corresponding protein were chosen as potent B-cell epitopes (Table 7 in Supplemental Data).
Final epitope selection and topological analysis of common epitopes
MHC Class I/II epitope prediction study and B-cell linear prediction were treated to find common epitopes. Common MHC class I-II epitopes with IC50 < 50 nM overlapped with B-cell predicted linear epitopes were shortlisted. 2 outer membrane proteins (Protein ID: Q7VF32, iron-regulated outer membrane protein FrpB; Protein ID: Q7VJ32, uncharacterized protein) and 3 extracellular proteins (Protein ID: Q7VH86, flagellar basal-body rod protein FlgG; Protein ID: Q7VJR0, flagellar basal body protein; Q7VJK1, Phage_base_V domain-containing protein) were finally selected based on their common T and B-cell epitopes (Table 4).
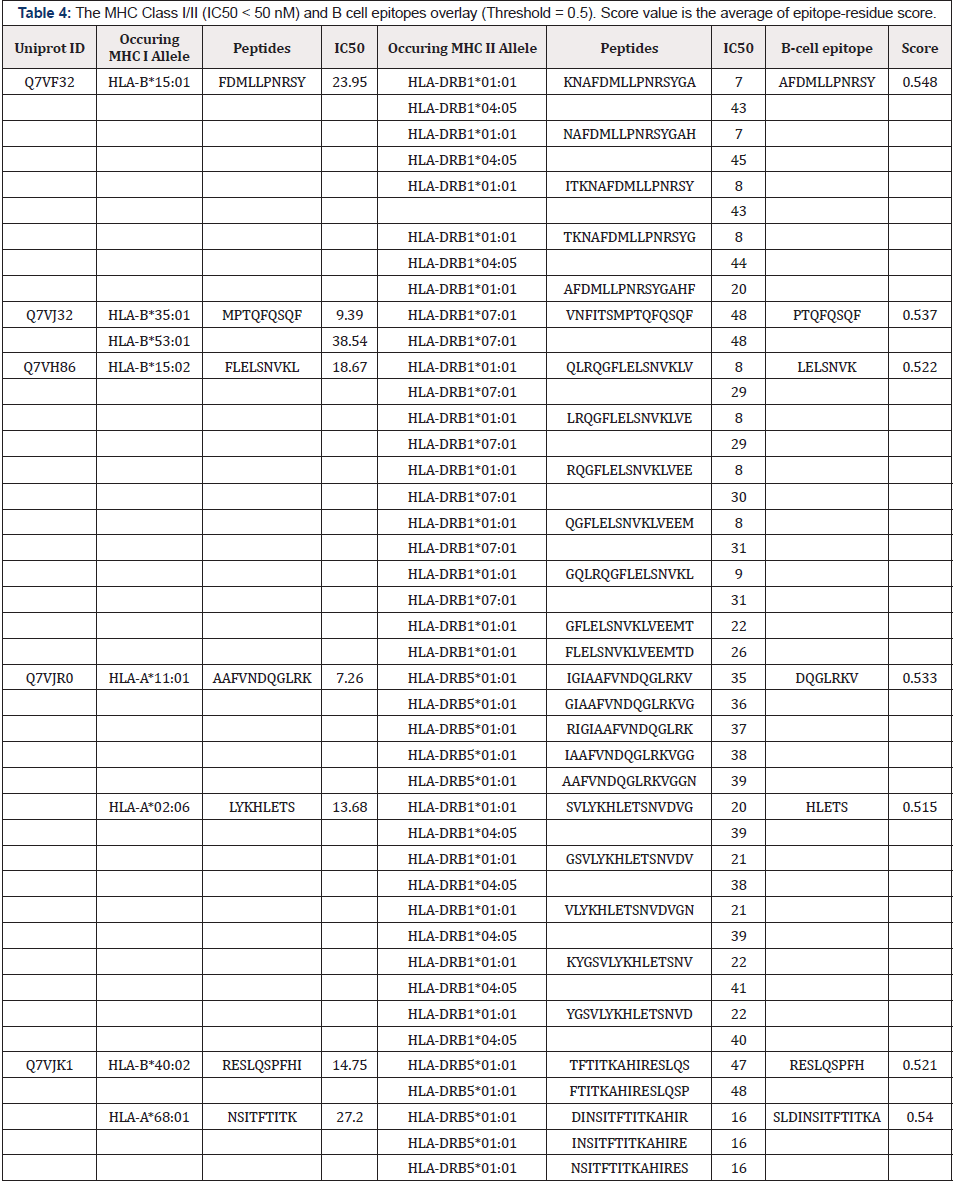
Table 4: The MHC Class I/II (IC50 < 50 nM) and B cell epitopes overlay (Threshold = 0.5). Score value is the average of epitope-residue score.
For the immune cells’ effective recognition, exo-topology of the predicted common epitopes was investigated. During the homology modeling, which was performed via SWISS-MODEL, 4AIP, 6JZR, 6K31, and 5FP1 were employed as templates of Q7VF32, Q7VH86, Q7VJR0, and Q7VJ32, respectively. Here templates were given as Protein Databank IDs (PDB), and target proteins were represented with UniProt IDs. In the case of Q7VJK1, which has two identified epitope regions from 10th to 27th amino acids, Phyre2 was employed in intense mode with the default settings to benefit from the ab-initio method because the templates identified for this protein represent a low coverage for the target region where the epitopes are located. After the analysis, the surface-exposed nature of the epitopes was displayed with no folding into the protein structure (Figure 4).
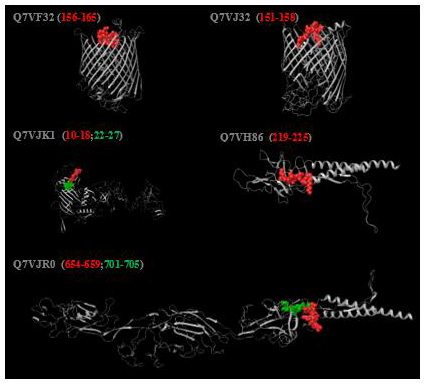
Figure 4: Conformational representation of the epitope regions on the structure of the related proteins. The protein structures are displayed in gray color and the epitopes are shown in red or green. Each of Q7VF32, Q7VJ32, and Q7VH68 proteins contains one epitope region within 156- 165 (FDMLLPNRSY), 151-158 (PTQFQSQF), and 219-225 (LELSNVK) bp, respectively. Q7VJK1 has two different epitope regions within 10-18 (NSITFTITK) and 22-27 (RESLQS) bp. Q7VJR0 also contains two epitope regions within 654-659 (DQGLRK) and 701-705 (HLETS).
Discussion
In the last decades, computational approaches, coupled with bioinformatics tools, have provided insights to identify potential targets and antigens from different microorganisms. The bioinformatics-mediated analysis helps narrow down the number of proteins or genes to be investigated through a rational workflow. This strategy provides advantages in terms of time and cost. Besides discovering novel targets or antigens, in-silico approaches can also help design structure-based drugs, repurpose known drugs, construct multi-epitope vaccine proteins, and investigate host-pathogen interactions [27]. A computational approach termed reverse vaccinology has been used to mine the genome or proteome of microorganisms to identify novel vaccine candidates, and sequence or structure-based comparisons with known targets provide information to discover putative drug targets.
In this study, a proteome-mediated analysis was applied to uncover potential antigenic proteins and drug targets in H. hepaticus, which is a member of the enterohepatic group of Helicobacter spp. with well-known pathogenesis in chronic hepatitis and hepatocellular carcinoma in mice. The studies also revealed its presence in human cases with cholecystitis, gallbladder cancer, liver diseases, and cholelithiasis, implying that a possible relationship between the infection of H. hepaticus and these diseases may be directly or indirectly present. After retrieving the total proteome of H. hepaticus, paralogous proteins or duplicates were eliminated through CD-HIT analysis. Human non-homologous proteins were also excluded since an ideal vaccine or drug candidate should not show homology with host proteins to prevent autoimmune reactions for vaccine candidates and avoid binding of therapeutic agents to the host homologs. Through the PSORTb and CELLO database, non-homologs were analyzed to identify the subcellular location, which is an important aspect since it helps to understand protein’s function and to evaluate proteins as drug targets or vaccine candidates.
As antibiotic resistance became a significant threat to human health, alternative approaches rather than antibiotics began to be sought to fight against bacterial pathogens. One popular approach is to design small-molecule enzyme inhibitors for critical pathways of microorganisms such as LPS, peptidoglycan, and quorum sensing [28]. In addition to traditional methods, genomics or proteomicsbased in-silico strategies enable to search potential druggable candidates for these small-molecule inhibitors in a fast manner [29-31]. Therefore, the cytoplasmic proteins of H. hepaticus were analyzed to reveal the enzymes that have a role in vital metabolic pathways that can serve as promising drug targets.
DEG 10 contains more than 12,000 essential genes and their expressed proteins from 31 bacterial species [16]. Besides search with the gene name or function, the database also allows BLASTp search for query sequences against essential genes. After retrieving essential gene-products among the cytoplasmic proteins through the DEG server, KEGG pathway analysis was carried out to reveal metabolic pathways unique to the pathogen. These analyses resulted in 36 proteins specific to 25 metabolic pathways found only in H. hepaticus compared to the human. Among them, LPS and peptidoglycan biosynthesis pathways have primary importance due to their critical roles, especially in pathogen survival, antibiotic sensitivity, and pathogenesis. Besides, the disruption of these constituents would be quite useful to control pathogens by making them vulnerable against osmotic lysis [26]. Therefore, fourteen out of thirty-six proteins assigned to these pathways were prioritized and analyzed, resulting in six hub proteins (UDP-N-acetylglucosamine acyltransferase (LpxA), UDP-3-O-acyl-N-acetylglucosamine deacetylase (LpxC), 2-dehydro-3-deoxyphosphooctonate aldolase (KdsA), 3-deoxymanno- octulosonate cytidylyltransferase (KdsB), phosphoheptose isomerase (GmhA) and penicillin-insensitive transglycosylase (PbpA)) whose inhibition would cause detrimental outcomes in the the pathogen’s survival (Table 2). In the literature, several studies show the importance of these indispensable enzymes in terms of the survivability of various bacterial pathogens [32-34].
LPS mainly consists of three distinct compartments: Lipid A, core oligosaccharide, and O-chain polysaccharide. Lipid A is a key element that anchors LPS in the outer part of the external membrane [35]. Among six putative drug targets, UDP-N-acetylglucosamine acyltransferase (LpxA) and UDP-3-O-acyl-N-acetylglucosamine deacetylase (LpxC) are the first and second enzymes, respectively, in the biosynthesis of Lipid A [36]. Many studies have been conducted in other pathogens to design small molecule or peptide inhibitors for these enzymes. Their blockage helped control the spread of the pathogens and decrease their pathogenicity. Besides, their combinational use with antibiotics made the cell wall weaker for the entrance of antibiotics into the host cells [37-40]. The second part, core oligosaccharide, is composed of a short chain of sugars such as KDO and heptose. 2-dehydro-3-deoxyphosphooctonate aldolase (KdsA) and 3-deoxy-manno-octulosonate cytidylyltransferase (KdsB) are the enzymes responsible for KDO biosynthesis. The studies carried out with different pathogens revealed that the inhibition of KdsA and KdsB resulted in the accumulation of Lipid A precursor, which eventually arrested the cell growth and made the bacterial cell more vulnerable to antibiotic actions [34,41]. In addition to KdsA and KdsB, phosphoheptose isomerase (GmhA) also has a role in the core oligosaccharide part where GmhA catalyzes the committed step of heptose biosynthesis, which in turn, could be a valuable target for the inhibition of bacterial growth [42]. The last enzyme, called penicillin-insensitive transglycosylase (PbpA), is a Class A penicillin-binding protein that catalyzes both transglycosylation and transpeptidation reactions in glycan strands of peptidoglycan. The studies have provided insights into its contribution to fully mature cell wall structure, viability, and resistance to β-lactam antibiotics [32]. When considering all these key enzymes’ functions, they seem to be valuable targets to control the pathogen due to the disruption of structural integrity and enabling antibiotics to accomplish their actions.
The outer membrane and extracellular proteins usually play significant roles as virulence factors related to the pathogen’s adhesion, invasion, and survival in the host. They are generally the first contact points with the host, which can elicit an immune response. Therefore, they can be considered as potential vaccine candidates, and Vaxign was used to analyze the outer membrane and extracellular proteins of H. hepaticus. Vaxign performs the topology analysis using HMMTOP based on a general hidden Markov model (HMM), and proteins having ≥ 2 transmembrane helices are not considered as ideal candidates due to the difficulty in the cloning and expression of proteins during vaccine studies [12,19]. Adhesin proteins are generally critical for pathogen invasion, and proteins with locations rather than cytoplasm and cytoplasmic membrane tend to have more adhesin probability (AP) [43]. Vaxign predicts AP using SPAAN with 89% sensitivity and 100% specificity. Moreover, Vaxign assigns a protegenicity score to each protein, indicating how well a candidate induces protective immunity [44]. After selecting the candidates based on Vaxign search, they were subjected to VFDB and VaxiJen analyses to screen virulent and antigenic proteins. VFDB contains up-todate virulence factors of various bacterial pathogens and supports BLASTp search based on sequence similarity. VaxiJen provides an alignment-independent prediction of antigenic proteins and makes antigen classification according to the physicochemical properties of proteins. The results exposed eleven putative vaccine candidates, including five uncharacterized proteins in addition to known immunogenic proteins; flagellar proteins and iron-regulated outer membrane protein (FrpB). Epitope analysis revealed that five of eleven vaccine candidates displayed common T and B-cell epitopes, which provide advantages to elicit both cellular and humoral immune responses (Table 4). Moreover, the structural analysis demonstrated the surface-exposure topology of these epitopes, suggesting an efficient immune system recognition. Among these five proteins, FrpB, FlgG, and flagellar basal-body protein were previously characterized, whereas two candidates remain uncharacterized. The immunogenic nature of bacterial flagella has been widely known, and flagellar proteins including FlaA, FlaB, FlgE, and FlgG studied in different pathogens have been found as immunogenic [45-48]. Another characterized protein, FrpB, has been also identified as immunogenic in various pathogens such as Neisseria meningitidis and Salmonella typhi [49, 50]. Besides these known antigens, Q7VJ32 (uncharacterized protein) and Q7VJK1 (phage base V domain-containing protein) could be considered as new antigenic determinants specific to H. hepaticus.
Conclusion
Although experimental validation is still needed, the current study uncovered six drug targets and eleven putative antigenic proteins in H. hepaticus through the subtractive proteomic approach. Given the fact that all six proteins have critical roles in vital metabolic pathways, they were typically identified as drug targets in different pathogens including Acinetobacter baumannii, Pseudomonas aeruginosa, and H. pylori in previous studies (Hamada et al., 2009; Pradhan and Dal, 2004; Rappuoli, 2000). Among eleven antigenic proteins, five had common epitopes of T and B-cells with an exo-topology, resulting in both humoral and cellular-mediated immune responses.
In further studies, structural analysis of the drug targets could be helpful to design specific small-molecule inhibitors to control the pathogen. Moreover, putative antigenic proteins could be used to detect H. hepaticus. The identified epitope regions can be utilized to speculate potential multi-epitope vaccine constructs with different adjuvants in further studies.
Acknowledgement
Not applicable.
Conflict of Interest
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The authors declare that they have no known competing personal relationships that could have appeared to influence the work reported in this paper.
References
- Viennois E, Gewirtz AT, Chassaing B (2019) Chronic inflammatory diseases: Are we ready for microbiota-based dietary intervention? Cell Mol Gastroenterol Hepatol 8(1): 61-71.
- Coussens LM, Werb Z (2010) Inflammation and cancer. Nature 420(6917): 860-867.
- Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, et al. (1994) Helicobacter hepaticus nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol 32(5): 1238-1245.
- Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH (2011) Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol 4(1): 22-30.
- Hamada T, Yokota K, Ayada K, Hirai K, Kamada T, et al. (2009) Detection of Helicobacter hepaticus in human bile samples of patients with biliary disease. Helicobacter 14(6): 545-551.
- Pradhan SB, Dal S (2004) Relation between gallbladder neoplasm and Helicobacter hepaticus Kathmandu Univ Med J (KUMJ) 2(4): 331-335.
- Nilsson I, Lindgren S, Eriksson S, Wadström T (2000) Serum antibodies to Helicobacter hepaticus and Helicobacter pylori in patients with chronic liver disease. Gut 46(3): 410-414.
- Shimoyama T, Takahashi R, Abe D, Mizuki I, Endo T, et al. (2010) Serological analysis of Helicobacter hepaticus infection in patients with biliary and pancreatic diseases. J Gastroenterol Hepatol 25(Suppl 1): S86-S89.
- Kleine M, Worbs T, Schrem H, Vondran FWR, Kaltenborn A, et al. (2014) Helicobacter hepaticus induces an inflammatory response in primary human hepatocytes. Plos One 9(6): e99713.
- Zhou D, Wang JD, Weng MZ, Zhang Y, Wang XF, et al. (2013) Infections of Helicobacter spp. in the biliary system are associated with biliary tract cancer: a meta-analysis. Eur J Gastroenterol Hepatol 25(4): 447-454.
- Rappuoli R (2000) Reverse vaccinology. Curr Opin Microbiol 3(5): 445-450.
- Naz A, Awan FM, Obaid A, Muhammad SA, Paracha RZ, et al. (2015) Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: A reverse vaccinology based approach. Infect Genet Evol 32: 280-291.
- Huang Y, Niu B, Gao Y, Fu L, Li W (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26(5): 680-682.
- Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64 (3): 643-651.
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, et al. (2010) PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26(13): 1608-1615.
- Luo H, Lin Y, Gao F, Zhang CT, Zhang R (2014) DEG 10, an update of the Database of Essential Genes that includes both protein-coding genes and non-coding genomic elements. Nucl Acids Res 42: D574-D580.
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6): 845-858.
- Hussein HA, Borrel A, Geneix C, Petitjean M, Regad L, et al. (2015) PockDrug-Server: a new web server for predicting pocket druggability on holo and apo proteins. Nucl Acids Res 43(W1): W436-W442.
- He Y, Xiang Z, Mobley HLT (2010) Vaxign: the first web-based vaccine design program for reverse vaccinology and an application for vaccine development. J Biomed Biotechnol 297505.
- Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumor antigens and subunit vaccines. BMC Bioinformatics 8: 4.
- Liu B, Zheng DD, Jin Q, Chen LH, Yang J (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucl Acids Res 47(D1): D687-D692.
- Solanki V, Tiwari V (2018) Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci Rep 8: 9044.
- Jespersen MC, Peters B, Nielsen M, Marcatilli P (2017) BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucl Acids Res 3(W1): W24-W29.
- Westbrook JD, Sho C, Feng Z, Zhuravleva M, Velankar S, et al. (2015) The chemical component dictionary, complete descriptions of constituent molecules in experimentally determined 3D macromolecules in the Protein Data Bank. Bioinformatics 31(8): 1274-1278.
- Ross K, Wu C, Damm W, Reboul M, Stevenson JM, et al. (2019) OPLS3e: extending force field coverage for drug-like small molecules. J Chem Theory Comput 15(3): 1863-1874.
- Aminemi U, Pradhan D, Marisetty H (2010) In silico identification of common putative drug targets in Leptospira interrogans. J Chem Biol 3(4): 165-173.
- Khan T, Mahmud A, Iqbal A, Hoque SF, Hasan M (2020) Substractive genomics approach towards the identification of novel therapeutic targets against human Bartonella bacilliformis. Inform Med Unlocked 20: 100385.
- Tomaras AP, McPherson CJ, Kuhn M, Carifa A, Mullins L, et al. (2014) LpxC inhibitors as new antibacterial agents and tools for studying regulation of lipid A biosynthesis in Gram-negative pathogens. mBio 5(5): e01551-e01514.
- Crowther GJ, Shanmugam D, Carmona SJ, Doyle MA, Hertz-Fowler C, et al. (2010) Identification of attractive drug targets in neglected-disease pathogens using an in silico approach. PLoS Negl Trop Dis 4(8): e804.
- Hassan SS, Jamal SB, Radusky LG, Tiwari S, Ullah A, et al. (2018) The druggable pocketome of Corynebacterium diphtheriae: a new approach for in silico putative druggable targets. Front Genet 9: 44.
- Froes TQ, Baldini RL, Vajda S, Castillho MS (2019) Structure-based druggability assessment of anti-virulence targets from Pseudomonas aeruginosa. Curr Protein Pept Sci 20(12): 1189.
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P (2008) The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32(2): 234-258.
- Sarkar M, Maganti L, Ghoshal N, Dutta C (2012) In silico quest for putative drug targets in Helicobacter pylori HPAG1: molecular modeling of candidate enzymes from lipopolysaccharide biosynthesis pathway. J Mol Model 18(5): 1855-1866.
- Ahmad S, Raza S, Abro A, Liedl KR, Azam SS (2019) Toward novel inhibitors against KdsB: a highly specific and selective broad-spectrum bacterial enzyme. J Biomol Struct Dyn 37(5): 1326-1345.
- Caroff M, Novikov A (2019) LPS structure, function, and heterogeneity. In: Williams K (Ed.), Endotoxin Detection and Control in Pharma, Limulus, and Mammalian Systems, Springer.
- Zhou P, Zhao J (2018) Structure, inhibition and regulation of essential Lipid A enzymes. Biochim Biophys Acta Mol Cell Biol Lipids 1862(11): 1424-1438.
- Dangkulwanich M, Raetz CRH, Williams AH (2019) Structure guided design of an antibacterial peptide that targets UDP-Nacetylglucosamine acyltransferase. Sci Rep 9(1): 3947.
- Jenkins RJ, Dotson GD (2012) Dual targeting antibacterial peptide inhibitor of early lipid A biosynthesis. ACS Chem Biol 7(7): 1170-1177.
- Tan JH, Vidaillac C, Yam JKH, Chua SL, Givskov M, et al. (2016) In vitro and in vivo efficacy of an LpxC inhibitor, CHIR-090, alone or combined with colistin against Pseudomonas aeruginosa biofilm. Antimicrob Agents Chemother 61(7): e02223-e02216.
- Garcia-Quintanilla M, Caro-Vega JM, Pulido MR, Moreno-Martinez P, Pachon J, et al. (2016) Inhibition of LpxC increases antibiotic susceptibility in Acinetobacter baumannii. Antimicrob Agent Chemother 60(8): 5076-5079.
- Pradhan D, Priyadarshini V, Munikumar M, Swargam S, Umamaheswari A (2013) 161 Discovery of potent KdsA inhibitors of Leptospira interrogans through homology modeling, docking, and molecular dynamics simulations. J Biomol Struct Dyn 31: 105.
- Umamaheswari A, Pradhan D, Hemanthkumar M (2010) Identification of potential Leptospira phosphoheptose isomerase inhibitors through virtual high-throughput screening. Genom Proteom Bioinf 8(4): 246-255.
- Ong E, Wong MU, He Y (2017) Identification of new features from known bacterial protective vaccine antigens enhances rational vaccine design. Front Immunol 8: 1382.
- Ong E, Wong MU, Huffman A, He Y (2020) COVID-19 Coronavirus vaccine design using reverse vaccinology and machine learning. Front Immunol 11: 1581.
- Wilhelm V, Miquel A, Burzio LO, Rosemblatt M, Engel E, et al. (2006) A vaccine against the salmonid pathogen Piscirickettsia salmonis based on recombinant proteins. Vaccine 24(23): 5083-5091.
- Jwang B, Dewing P, Fikrig E, Flavell RA (1995) The hook protein of Borrelia burgdorferi, encoded by the flgE gene, is serologically recognized in Lyme disease. Clin Dian Lab Immunol 2(5): 609-615.
- Yamazaki K, Kashimoto T, Hashimoto Y, Kado T, Ueno S (2018) Immunogenicity and protective efficacy of Vibrio vulnificus flagellin protein FlaB in a wound infection model. J Vet Med Sci 80(1): 55-58.
- Zarei M, Mosayebi G, Khansarinejad B, Abhati H (2017) Antigenic and immunogenic evaluation of Helicobacter pylori FlaA epitopes. Iran J Basic Med Sci 20(8): 920-926.
- Ala’Aldeen DA, Davies HA, Borriello SP (1994) Vaccine potential of meningococcal FrpB: studies on surface exposure and functional attributes of common epitopes. Vaccine 12(6): 535-541.
- Sood S, Rishi P, Dhawan V, Sharma S, Ganguly NK (2005) Protection mediated by antibodies to iron-regulated outer-membrane proteins of S. typhi in a mouse peritonitis model. Mol Cell Biochem 273(1-2): 69-78.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.