Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Identification Small Molecules Inhibit the SARS-Cov-2 Spike-ACE2 (PPI)
*Corresponding author: Yara mansour, Faculty of Pharmacy, Helwan University, Ain-Helwan, Egypt
Received: June 01, 2021; Published: June 09, 2021
DOI: 10.34297/AJBSR.2021.12.001838
Keywords: (hACE2), Angiotensin-Converting Enzyme-2 (ACE2), Peptides, Small molecules, Receptor-Binding Domain (RBD), Immunogenicity, Proteins, Solubility, Unsuitability, Drug, Inhibition PPI, Disruptors
Introduction
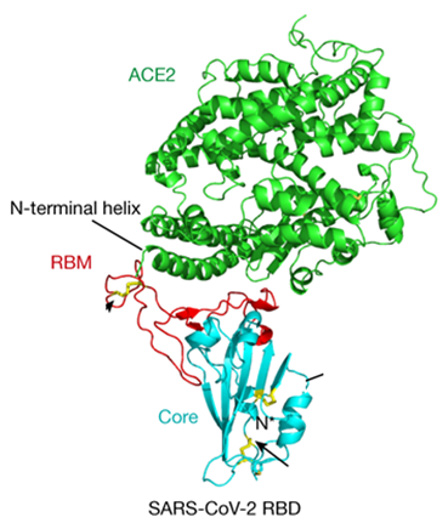
Acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) is responsible for COVID-19 which reached pandemic levels in early 2020 (WHO; March 11, 2020) [1,2]. There are three types of CoVs that reached pandemic levels in the last twenty years: SARS-CoV-1, MERS-CoV and SARS-CoV-2 [3]. Coronaviruses use the Receptor- Binding Domain (RBD) of the homotrimeric spike (s) glycoprotein emerging from its surface of the envelope to bind to their cellular receptors [4]. This binding facilitates a cascade of events that leads to the penetration host cell and finally the virus replication [5]. Binding to the Angiotensin-Converting Enzyme-2 (ACE2) receptor is a key consideration step for entering SARS-CoV into target cells [6-9]. The Protein-Protein Interaction [10] (PPI) between the SARS-CoV-2 spike protein and Human ACE2 (hACE2) (Figure1), in the early stages of the life cycle created a significant therapeutic need for possible antiviral drug development. The current SARSCoV antibodies are not effective in case SARS-CoV-2 [11]. All protein therapies showed great problems such as solubility, unsuitability for oral or inhaled administration, and immunogenicity. In addition, being foreign proteins in nature work as antigens and elicit strong immune responses in certain patients [12].
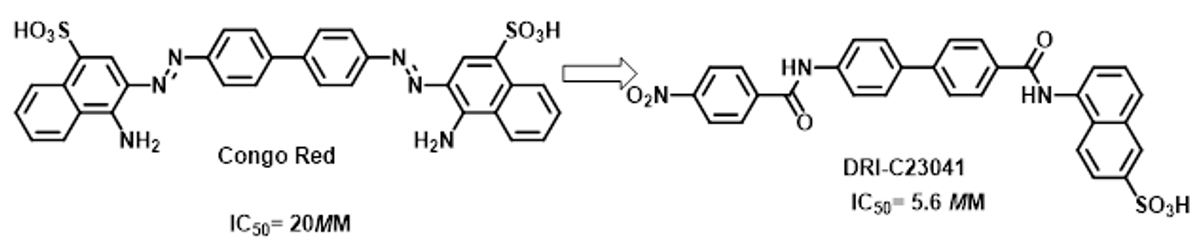
Peptides or small molecules demonstrate promising approaches. There are some reported peptides as PPI disruptors, but so far none have been very effective [13-15]. Lack of welldefined binding pockets on the protein surface that would allow their adequate binding to decrease the possibilities for successful small molecules inhibitors. Buchwald [16] and his team, identify new compounds that inhibit PPI at low micromolar levels by screening small molecule libraries such as Congo Red and Evans Blue. Theses organic dyes showed excellent protein binders and its structures demonstrate that designed as such, that is supporting a drug discovery research. The dye itself is usually well known lack therapeutic effect for its pigmentation, and rapidly decompose in case azo dyes. There are overlap between the existing chemical space of drug like structures and inhibition PPI. Obviously, it clarified the structural motif for active compounds: a biphenyl linker with a naphthyl at one end and another aromatic naphthyl or phenyl at the other end, both with at least one polar substituent. Importantly, these criteria support identification of novel and promising SMIs that are more druglike, free of color-causing chromophore as well as low micromolar activity in cell-free ELISAtype assays16 (Figure2).
References
- JB Moore, CH June (2020) Cytokine release syndrome in severe COVID-19. Science 368(6490): 473-474.
- NJ Matheson, PJ Lehner (2020) How does SARS-CoV-2 cause COVID-19? Science 369(6503): 510-511.
- DD Rajgor, MH Lee, S Archuleta, N Bagdasarian, SC Quek, (2020) The many estimates of the COVID-19 case fatality rate. Lancet Infect Dis 20(7): 776-777.
- D Wrapp, N Wang, KS Corbett, JA Goldsmith, CL Hsieh, et al. (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367(6483): 1260-1263.
- H Sivaraman, SY Er, YK Choong, E Gavor, J Sivaraman, (2021) Structural basis of the SARS-CoV-2/SARS-CoV receptor binding and small-molecule blockers as potential therapeutics. Annu Rev Pharmacol Toxicol 6(61): 465-493.
- P Zhou, Xing Lou Y, Xian Guang W, Ben Hu, Lei Zhang, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798): 270-273.
- Walls, AC, Young Jun P, M Alejandra, Abigail Wall, McGuire AT, et al. (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2): 281-292.
- M Letko, A Marzi, V Munster (2020) Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B beta coronaviruses. Nat Microbiol 5(4): 562-569.
- M Hoffmann, H Kleine Weber, S Schroeder, N Kruger, T Herrler, et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2): 271-280.
- J Lan, J Ge, J Yu, S Shan, H Zhou, et al. (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807): 215-220.
- S Xiu, A Dick, H Ju, S Mirzaie, F Abdi, et al. (2020) Inhibitors of SARS-CoV-2 entry: current and future opportunities. J Med Chem 63 (21): 12256-12274.
- B Leader, QJ Baca, DE Golan, (2008) Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discovery 7(1): 21-39.
- S Xiu, A Dick, H Ju, S Mirzaie, F Abdi, et al. (2020) Inhibitors of SARS-CoV-2 entry: current and future opportunities. J Med Chem 63(21): 2256-12274.
- C Gil, T Ginex, I Maestro, V Nozal, L Barrado Gil, et al. (2020) COVID-19: Drug targets and potential treatments. J Med Chem 63(21): 12359-12386.
- G Zhang, S Pomplun, AR Loftis, X Tan, A Loas, et al. (2020) Investigation of ACE2 N-terminal fragments binding to SARS-CoV-2 Spike RBD. bioRxiv 20: 1.
- D Bojadzic, O Alcazar, J Chen, S Chuang, JM Condor Capcha, et al. (2021) Small-Molecule Inhibitors of the Coronavirus Spike: ACE2 Protein-Protein Interaction as Blockers of Viral Attachment and Entry for SARS-CoV‑ ACS Infect Dis XXX: XXX-XXX



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.