Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Recombinant Protein of Immunogenic Metabolic Enzyme Epitopes of Trichomonas vaginalis are Common to Humans and Microorganisms
*Corresponding author: JF Alderete, School of Molecular Biosciences, College of Veterinary Medicine, Washington State University, Pullman, Washington, USA
Received: July 20, 2021; Published: August 17, 2021
DOI: 10.34297/AJBSR.2021.13.001930
The number one, non-viral sexually transmitted infection (STI) worldwide is caused by the ancient protist Trichomonas vaginalis. Recently, I reported on an ~40-kDa String-Of-Epitopes chimeric protein (AEG::SOE2) consisting of immunogenic epitopes unique to the Trichomonas vaginalis fructose-1,6bisphosphate aldolase (A), α-enolase (E), and glyceraldehyde-3-phosphate dehydrogenase (G). This report is on another construct of a 49.41-kDa AEG::SOE3 chimeric protein comprised of 8, 9 and 12 non-unique, immunogenic epitopes of A, E and G, respectively. These non-unique epitopes were detected by sera of women and men patients but not control, seronegative sera of women and men. The epitopes were found to have ≥50% to 100% sequence amino acid sequence identity with the human homolog and homologs of Candida albicans, Escherichia coli, Saccharomyces cerevisiae, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes as well as the homologs of the STI agents Chlamydia trachomatis, Neisseria gonorrhoeae and Treponema pallidum. The AEG::SOE3 protein was not detected by mouse anti AEG::SOE2 serum and by monoclonal antibodies (MAbs) to AEG::SOE2, showing the distinctness of these two chimeric proteins. AEG::SOE3 was detected by ELISA and immunoblot with sera of mice immunized with fixed T. vaginalis organisms, with sera of women and men patients with trichomonosis, and with MAbs to AEG::SOE3. The patient sera reactive to these immunogenic metabolic enzyme epitopes of AEG::SOE3 may be indicative of immune-crossreactive antibodies to human proteins during this STI. The role of these possible immune-crossreactive antibodies produced during this STI and those conceivably generated by other STIs and microbial pathogens to heretofore unappreciated disease pathogenesis is discussed.
Keywords: Candida Albicans; Chlamydia Trachomatis; Diagnostic, Diagnostic Targets; Enzyme Linked Immunosorbent Assay (ELISA); Epitope Chain Protein; Neisseria Gonorrhoeae; Non-Unique Epitopes, Immunogens; Point-Of-Care (POC); Streptococcus Pneumoniae; Streptococcus Pyogenes; Sera; Serodiagnosis; Sexually Transmitted Infections (Stis); Treponema Pallidum; Trichomonas Vaginalis; Unique Epitopes
Abbreviations: A: fructose-1,6-bisphophate aldolase; E: α enolase; ELISA: Enzyme-linked immunosorbent assay; G: glyceraldehyde-3-phosphate dehydrogenase; MAb: monoclonal antibody; POC: Point-of-Care; SDSPAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; STIs: Sexually Transmitted Infections; SOE: String-of-Epitopes
Introduction
Trichomonas vaginalis is responsible for the number one, non-viral STI that negatively affects women’s reproductive health [1,2]. It is evident that the global prevalence of this and other STIs requires specific and sensitive point-of-care (POC) diagnostics suitable for screening large cohorts of at-risk individuals. This is especially the case at low resource settings. A POC Trichomonas Rapid Test (Sekisui Diagnostics, Lexington, MA, USA) for diagnosing trichomonosis in women was invented in my laboratory [3], but it requires a vaginal swab from women and cannot diagnose the STI in men. Recent work has demonstrated two perfect targets [4,5] that can either be used singly or in combination for a POC serodiagnostic test useful for resource-poor environments worldwide. In a recent paper [4] a chimeric String-of-Epitopes (SOE) recombinant protein called AEG::SOE2 was shown to be a specific serodiagnostic target for this STI. This protein has been referred to as an epitope chain protein [6] that may have applicability as a vaccine. This protein has immunogenic epitopes of fructose-1,6-bisphosphate aldolase (A), α-enolase (E), and glyceraldehyde-3-phosphate dehydrogenase (G) that are unique to the T. vaginalis proteins [4]. A second recombinant protein target for a serodiagnostic test, referred to as ACT::SOE3, is a 72.4-kDa truncated version of the 106.2-kDa highly immunogenic α actinin protein to which women and men patients make serum IgG antibodies [7]. This protein is also unique to T. vaginalis in that it has no sequence identity with other proteins in databanks. This protein target has seven epitopes detected by women, within which are five epitopes reactive with men patients [5,7]. Both the AEG:: SOE2 and ACT::SOE3 serodiagnostics have utility for large-scale screening for this STI, which is preparatory to advancing the reproductive health of at-risk humans.
In the earlier paper on AEG::SOE2 [4] it became evident that A, E, and G also had immunogenic epitopes that were not unique to T. vaginalis. In this report another novel recombinant chimeric protein called AEG:: SOE3 is presented. This epitope chain protein is comprised of the non-unique epitopes of A, E and G that have ≥50% to 100% identity with homologous proteins of humans and of Candida albicans, Escherichia coli, Saccharomyces cerevisiae, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, including Chlamydia trachomatis, Neisseria gonorrhoeae, and Treponema pallidum, which also cause STIs [8]. Based on numerous published papers [9-18], I hypothesize that these non-unique, immunogenic epitopes with possible immuno-crossreactivity with human proteins may play a role in disease pathogenesis during this STI. I further discuss and hypothesize that T. vaginalis, other STIs and infectious diseases may produce immune-crossreactive antibodies during natural infections, and these antibodies may cause previously unreported and unappreciated host cytopathology.
Methods
Non-unique epitopes to the A, E and G proteins of T. vaginalis
The identification of the immunogenic epitopes of A, E and G reactive with sera of women and men patients was performed using the SPOTS system (Sigma-Genosys, The Woodlands, TX, USA) as detailed in several earlier reports [4,5,7,8]. Briefly, oligopeptides were derived from GenBank® accession numbers AAW78361, AAK73099 and AAA30325 for sequences of T. vaginalis of A, E and G proteins, respectively. Commercially available oligopeptides (Custom Peptide Arrays) were synthesized on activated membranes on the SPOTS system for screening with patient sera of women and men using established [4,5,7,8] and the manufacturer’s protocols. The membranes were also screened with monoclonal antibodies (MAbs) generated to the A, E and G proteins, as described below and previously [4,8]. The BLAST sequence identity analysis permitted identification of the epitopes (Table 1) with ≥50% to 100% amino acid sequence identity to other enzyme homologs, as before [8]. Finally, and as recently reported [4], the Immune Epitope Database and Analysis Resource (www.iedb.org) was used to show the linear nature of the epitope sequences.
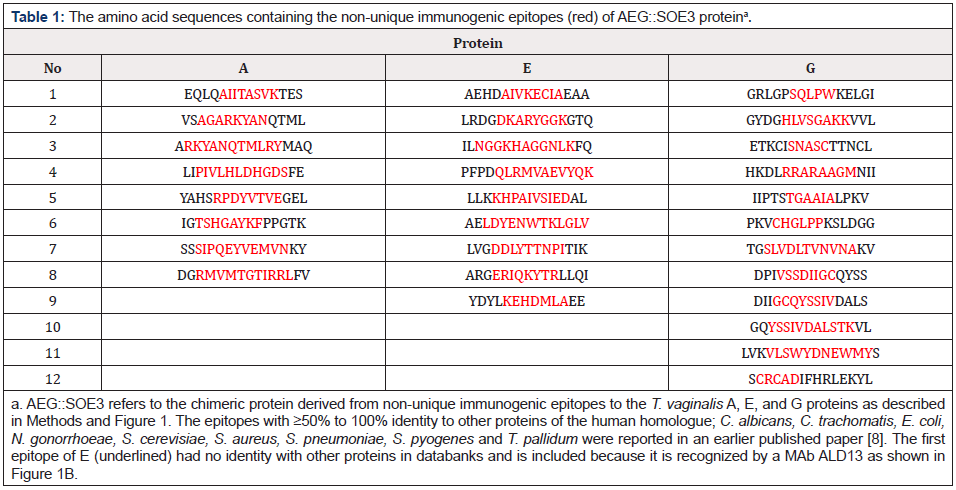
Table 1: The amino acid sequences containing the non-unique immunogenic epitopes (red) of AEG::SOE3 proteina.
Plasmid encoding AEG::SOE3 and purification of the protein
GenWay Biotech, Inc (San Diego, CA) synthesized the DNA coding sequence for AEG::SOE3 in the pET23a(+) plasmid with ampicillin and chloramphenicol genes. The protein as shown in Figure 1 was synthesized by induced E. coli BL21DE3 cells, as before [4,5,7,8]. AEG::SOE3 has hexa-histidine at the carboxy terminus for purification by Ni-NTA superflow affinity column chromatography (Qiagen Inc., Valencia, CA USA) [4]. The amino acid sequence of the protein was confirmed by sequencing of the plasmid. Because the protein was found within inclusion bodies, GenWay provided the purified protein for this study. Alternatively, the protein was purified in the laboratory by growing E. coli on Luria Broth with the antibiotics as described recently [4,5,7]. Briefly, a starter culture of 100ml of E. coli was grown at 37°C to OD600. The E. coli was then transferred to a 2-liter Erlenmeyer flask with 600ml Luria Broth and placed in a shaking incubator at 18°C prior for 2-hours prior to induction with isopropyl-β-Dthiogalactopyranoside and incubated for 4-hours. The protein was purified in the Ni-NTA column as shown in (Figure 2).

Figure 1: A. Linear amino acid sequence of the AEG::SOE3 protein of 436 amino acids with the hexa-His tag at the carboxy terminal end. The peptide-epitope sequences are linked by EE amino acids (underlined). Epitopes within the peptide sequences are red. B. Linear sequence with A, E and G refer to amino acid sequences from the T. vaginalis proteins, respectively, as described for Table 1. The epitopes were detected by pooled positive women (W) or men (M) sera, as shown and as indicated previously [8], and the number next to W and M are the order the epitope was identified within the protein during epitope mapping on the SPOTs system (Methods). The MAbs labeled ALD11, ALD12, ALD13, ALD25, B44 and B43 are reactive with the epitopes, as before [8].
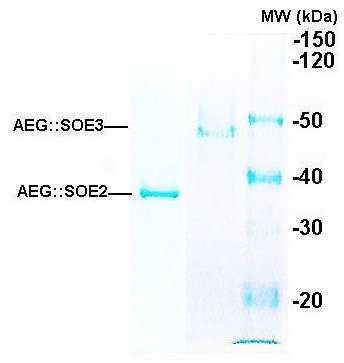
Figure 2: Representative SDS-PAGE in 8% acrylamide of the 35.9-kDa AEG::SOE2 protein (left lane) as recently reported [4] and the 49.4-kDa AEG::SOE3 protein (middle lane). Both proteins were purified by Ni2+NTA affinity chromatography, as before [4] (Methods). The right lane shows the molecular weight (MW) standards, and numbers refer to daltons (kDa, x1000).
Enzyme-linked immunosorbent assay (ELISA)
The preparation of the microtiter plates with AEG::SOE3 for ELISA is as before [4,5,7,8]. For all assays 2% ELISA-grade bovine serum albumin (eBSA) (Sigma Chemical Co., St. Louis, MO, USA) prepared in PBS (eBSA-PBS) was used for blocking the wells of microtiter plates and for dilution of sera. The eBSA-PBS also served as a negative control during ELISAs (Table 2). As shown in Figure 2,3 in addition to negative controls, ELISA was performed using recently generated mouse anti-AEG::SOE2 and MAbs to AEG:: SOE2 [4], mouse anti-T. vaginalis (Tv) serum [19], and three different MAb cocktails to AEG::SOE3 (described below), and pooled sera of women and men patients, as before [4,5,7,8].
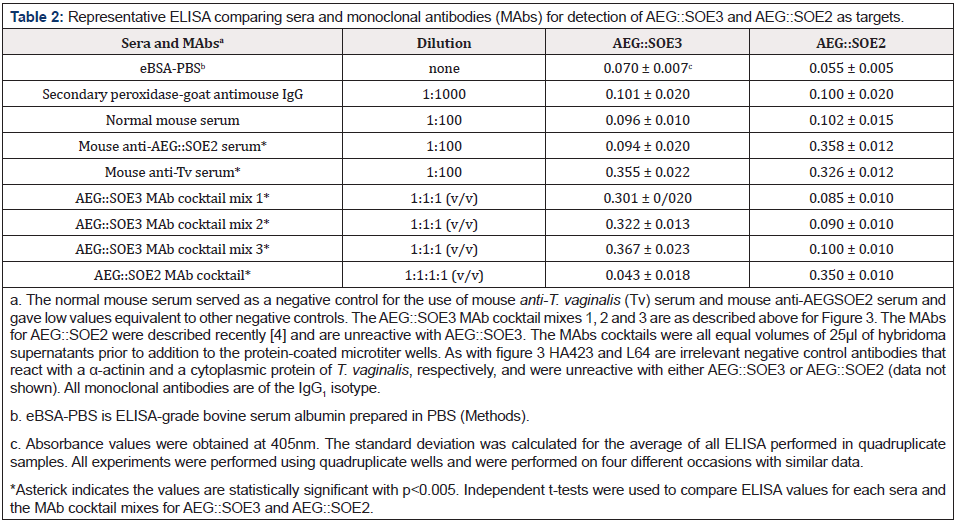
Table 2: Representative ELISA comparing sera and monoclonal antibodies (MAbs) for detection of AEG::SOE3 and AEG::SOE2 as targets.
Antibodies for detection of proteins and trichomonads
Numerous reports describe the sera from women and men used for detection of proteins to T. vaginalis [4,5,7,8]. These sera permitted determining levels of serum IgG antibodies to trichomonad proteins and particularly α-actinin [5]. Individual seropositive sera of women and men had identical reactivities to parasite proteins, and the limitation of amounts of individual sera was overcome by pooling of the seropositive sera of both women and men to obtain sufficient volumes for doing epitope mapping analysis. Likewise, pooled negative control sera from both women and men were used in ELISA experiments. Institutional Review Board (IRB) approvals for using the sera were obtained. A 1:25 (Vol: Vol) dilution of sera with eBSA-PBS was previously found to be optimal for detection of purified proteins immobilized onto wells of microtiter plates [4,5,7,8]. When doing ELISAs using MAbs to AEG::SOE3 (Figure 1B), a 50µl volume of undiluted hybridoma supernatant was added to wells. Alternatively, a cocktail of 25µl of hybridoma supernatant for each of MAb cocktails was used as presented (Figure2,3).
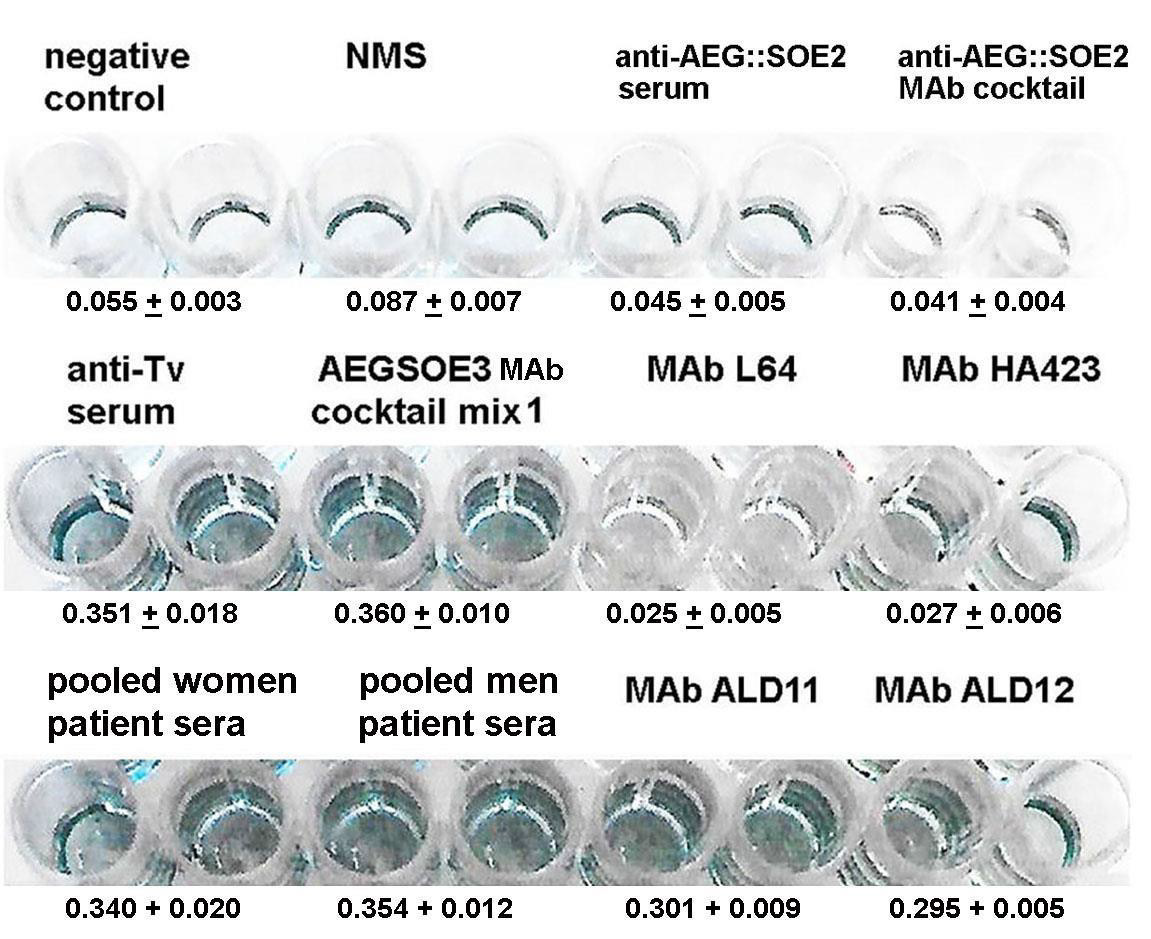
Figure 3: Representative ELISA for detection of 1 μg AEG::SOE3 immobilized onto individual wells of 96-well microtiter plates (Methods). The negative control is the secondary peroxidase-conjugated goat anti-mouse IgG. Normal mouse serum (NMS), mouse anti-AEG::SOE2 serum and mouse anti-T. vaginalis (Tv) serum were each diluted 1:100 in eBSA-PBS. The mouse anti-AEG::SOE2 serum was described recently [4] and was unreactive with AEG::SOE3. Mouse anti-T. vaginalis (Tv) serum was generated to chemically stabilized T. vaginalis isolate NYH 286 [19]. The AEG::SOE3 cocktail mix 1 was comprised of the MAbs ALD12, ALD11, ALD25. Two other MAb cocktails included mix 2 with ALD12, B44, B43 and mix 3 with ALD12, ALD11, ALD25, B44, B43 and gave results as mix 1. The MAbs were to epitopes as per Figure 1B. The MAb cocktails included 25μl of each MAb. The AEG::SOE2 MAb cocktail as recently reported [4] consisted of MAbs ALD13, ALD55, ALD30 and ALD32 and did not detect AEG::SOE3. The individual MAbs ALD11 and ALD12 detected AEG::SOE3. As expected, negative control irrelevant MAbs HA423 to α-actinin and L64 to a cytoplasmic protein [4,5,7,8] of T. vaginalis were unreactive to AEG::SOE3. All MAbs were of the IgG1 isotype. Women and men patient sera have been described before, and the optimal pooled sera dilution was 1:25 (vol/vol) in eBSA-PBS [4,5,7,8]. The ELISA was repeated on four different occasions using quadruplicate samples and gave similar data.
Mouse anti-AEG::SOE2 serum was used and described before [4]. Further, mouse anti-T. vaginalis serum generated in mice immunized with glutaraldehyde-fixed trichomonads has been reported [19]. The MAbs to A, E and G proteins were generated at the MAB Core Facility of the College of Veterinary Medicine at Washington State University using established protocols described before [7]. All animals were handled humanely as per guidelines by the Institutional Animal Care and Use Committee (IACUC number 6317) and National Institutes of Health protocols. After establishing that mouse anti-serum had antibody reactive to the three proteins, hybridomas were generated and used in the SPOT system for identifying the epitope amino acid sequences for each MAb. All MAbs are of the IgG1 isotype. The new MAbs (ALD11, ALD12, ALD13, ALD25) and the MAbs B44 and B43 used in earlier studies [8,20,21] were analyzed further with AEG::SOE3.
Statistical analysis
All statistical analyses were conducted in R (Version 4.0.4). Independent t-tests were used to examine differences in ELISA results between AEG::SOE3 and AEG::SOE2 for each sera and MAbs examined. Statistical significance was defined as a two-sided p-value less than 0.05.
Reproducibility
Experiments were performed on at least four occasions using quadruplicate samples under identical conditions and gave similar data. Averages and standard deviations were calculated.
Results
The T. vaginalis non-unique, immunogenic epitopes of A, E, and G and the AEG::SOE3 protein
An earlier report [8] on epitope mapping revealed that pooled sera of women and men patients recognized 12 epitopes of fructose-1,6-bisphosphate aldolase (A), 18 epitopes of α-enolase (E), and 19 epitopes of glyceraldehyde-3-phosphate dehydrogenase (G) for a total of 49 epitopes for the three proteins [18]. Eighteen of these 49 epitopes were found to be unique to the A, E and G T. vaginalis proteins [4,8]. In this report Table 1 shows the non-unique epitopes (colored red) identified by IgG antibody in the SPOTS system (Methods). There were 8 epitopes for A, 8 epitopes for E, and 12 epitopes for G for a total of 28 non-unique epitopes to these proteins. Except for the first epitope for E, these epitope amino acid sequences had ≥50% to 100% identity with other bacterial, fungal, and human sequences [8]. The first sequence for E (underlined) had no identity with other proteins in databanks and is included as an internal control because the epitope is recognized by MAb ALD13 (Figure 1).
AEG::SOE3 protein sequence
Figure 1A presents the recombinant protein amino acid sequence of the peptides containing the 15mer sequences shown in Table 1. Each peptide was linked with two glutamic acid residues (underlined). The protein with six histidine residues at the carboxy terminus is 436 amino acids in length and is 49.41kDa with a pI of 4.83. Figure 1B shows the reactivity with women (W) and/or men (M) positive sera indicated above the red epitope amino acid sequences. The epitopes for A(M1) and A(W2) overlapped. Overall, there are 17 and 14 epitopes detected by women and men sera, respectively. The six monoclonal antibodies (MAbs) that detect epitopes are labeled ALD11, ALD12, ALD13, ALD25, B44 and B43, as indicated. These MAbs have been previously characterized [8].
Purification of recombinant AEG::SOE3
Figure 2 shows the Coomassie Brilliant blue-stained gel after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDSPAGE) of purified AEG::SOE2 (left lane) and AEG::SOE3 (middle lane) after Ni-NTA affinity chromatography. The relative mobility of the protein was compared with molecular weight standards (right lane), and the proteins are consistent with the expected size of ~35.9-kDa for AEG::SOE2 and 49.41-kDa for AEG::SOE3.
ELISA using AEG::SOE3-coated wells probed with different antibodies
ELISA was then performed with 1 μg AEG::SOE3 immobilized onto microtiter wells, as before [4], and representative results of four separate experiments, each with quadruplicate samples, are shown in Figure 3. All experiments yielded similar data. One microgram of protein was found to be optimal for detection by antibody [4,5,7,8]. Not unexpectedly, there was no reaction of AEG::SOE3 by the secondary peroxidaseconjugated goat anti-mouse IgG (negative control) and normal mouse serum (NMS). Both the mouse antiAEG::SOE2 serum and a cocktail of MAbs to AEG::SOE2 previously reported [4,8] were unreactive with AEG::SOE3. The protein was detected by mouse anti-T. vaginalis serum (anti-Tv serum) diluted 1:100, an AEG::SOE3 MAb cocktail mix 1 (ALD12, ALD11, ALD25), and the individual MAbs ALD 11 and ALD12. Not shown is that two other cocktails of MAbs (mix 2 of ALD12, B44, B43 and mix 3 of ALD12, ALD11, ALD25, B44, B43) also gave identical results as shown for mix 1. The AEG::SOE3 recombinant protein shown in Figure 2 was detected by immunoblot with these antibody reagents and pooled sera of women and men patients (not shown) as was done for AEG::SOE2 recently [4]. Pooled patient sera from 5 women and 5 men readily detected AEG::SOE3. The α-actinin protein MAb HA423 and a cytoplasmic protein MAb L64 of T. vaginalis [4,5,7,8] were other negative controls and unreactive to AEG::SOE3. All MAbs were of the IgG1 isotype. Statistical analysis (Methods) was performed to compare ELISA reactions with sera and MAbs to AEG::SOE2 and AEG::SOE3 that the values were statistically significant as shown in Table 2.
Representative ELISA comparing ACT::SOE3 and AEG::SOE2 with different antibodies
AEG::SOE3 was then compared with AEG::SOE2 for detection by IgG of different antibodies. Table 2 presents results from a representative experiment with quadruplicate samples that was repeated four different times, and each experiment gave similar data. The negative controls included the eBSA-PBS alone, secondary peroxidase-conjugated goat anti-mouse IgG and normal mouse serum. Additional negative controls not shown include the irrelevant MAbs HA423 to α-actinin and L64 to a cytoplasmic protein [4], as shown above in Figure 3. As expected, the mouse anti- AEG::SOE2 serum used before [4] was reactive with AEG::SOE2 but not with AEG::SOE3. The mouse anti-T. vaginalis (Tv) serum readily detected both AEG::SOE3 and AEG::SOE2 recombinant proteins. Not surprisingly, three different cocktails of MAbs to AEG::SOE3 as indicated above (Figure 3) readily detected AEG::SOE3 but not AEG::SOE2. As expected, a MAb cocktail to AEG::SOE2 [4] readily detected AEG::SOE2 but was unreactive with AEG::SOE3. The asterick indicates that the values comparing ELISA reactions for AEG::SOE3 and AEG::SOE2 were statistically significant defined as a two-sided p values less than 0.05. Collectively, these data show the distinctness of the two recombinant proteins.
Discussion
The approach recently published [4] was used to synthesize another epitope chain protein [6] called AEG::SOE3 (Figures 1,2). This AEG::SOE3 is different from AEG::SOE2 because the epitopes of fructose1,6-bisphosphate aldolase (A), α-enolase (E), and glyceraldehyde-3-phosphate dehydrogenase (G) are not unique to T. vaginalis and share ≥50% to 100% amino acid identity with the human homologue as well as homologues of C. albicans, E. coli, S. cerevisiae, S. aureus, S. pneumoniae, and S. pyogenes and the three other STI agents, C. trachomatis, N. gonorrhoeae, and T. pallidum [8]. The fact that mouse anti-AEG::SOE2 sera and MAbs to AEG::SOE2 [4] were unreactive with AEG::SOE3 and that MAbs to AEG::SOE3 did not detect AEG::SOE2 shows the specific and distinct nature of the different epitopes and the two proteins. Further, data from the earlier work [4] and in this study support the surface location of the epitopes of these metabolic enzymes. For example, mouse antiserum generated to chemically-stabilized T. vaginalis [19] reacts to both AEG::SOE2 [4] and AEG::SOE3 (Figure 2,3). In addition, it was shown recently that this antiserum and MAbs detects trichomonads in a whole-cell ELISA [4]. These data and earlier reports [20,21] show that the enzymes E and G reside on the surface of T. vaginalis. These surface-associated enzymes are ligands for binding host pathogenicity proteins, like plasminogen and other proteins, such as fibronectin, laminin, and collagen [20,21]. That there is an antibody response to these epitopes during infection by T. vaginalis may indicate that the host immune surveillance recognizes these enzyme proteins not as homologs of human proteins but as virulence factors involved in disease pathogenesis.
It is noteworthy that the host immune response resulted in IgG antibodies to 12 immunogenic epitopes of A, 18 epitopes of E and 19 epitopes of G given the highly conserved nature of the proteins among humans. Of these 49 total epitopes, 8 of 12 epitopes of A, 9 of 18 epitopes of E and 12 of 19 epitopes of G were not unique to T. vaginalis and have ≥50% to 100% sequence identity with homologues of humans and other bacterial pathogens and fungi, including three other organisms that cause STIs. The AEG::SOE3 epitopes with high sequence identity to C. trachomatis, N. gonorrhoeae and T. pallidum enzymes perhaps suggests that IgG antibodies may be made to these enzyme epitopes during infection by the other STI agents as well. If this is true, this fact would be significant and may further suggest that assumptions that the immune response to microbial pathogens may not aggressively be towards protein homologues of humans requires additional attention.
It is important to consider that antibodies to these non-unique trichomonad enzyme epitopes may elicit immune cross-reactions with host proteins and tissues. While it is acknowledged that this requires future examination, at a minimum this work suggests that this is an issue that should be considered within the framework of pathogenesis of trichomonosis, especially as it has been shown that human serum antibody to α–enolase of group A streptococcus cross-reacts with host tissues [22]. Indeed, there is a growing literature showing autoantibody in relation to infectious diseases [9-18]. For example, bacterial pathogens, viruses, and parasites are now considered triggers of autoimmunity [9,10]. Hepatitis C in patients is considered in cases of autoimmune encephalitis due to West Nile virus and other bacteria [11,18]. The role of viruses and microbial pathogens is related to multiple sclerosis [9]. Among some organisms implicated in diseases are the following: Chlamydia pneumoniae and biliary cirrhosis [15], Helicobacter pylori and autoimmune gastritis [12], Trypanosoma cruzi and Chagas’ cardiomyopathy [13], Leishmania and autoimmune hepatitis [17] and Mycoplasma arthritidis and rheumatoid arthritis [16]. Therefore, it is reasonable to hypothesize that the epitopes presented in Table 1 with sequence identities to human and other microbial pathogen proteins may play a role in heretofore unknown and unappreciated disease pathogenesis via eliciting host immuno-crossreactive antibodies.
These findings strongly reaffirm an earlier report [4] that argues against the use of whole cells and/or lysates as diagnostics and vaccines [23-25]. The ideal serodiagnostic target would be a protein with immunogenic epitopes unique to the pathogen of interest [4]. It is shown here that A, E and G of T. vaginalis leads to IgG to epitopes shared with other infectious diseases, including C. trachomatis, N. gonorrhoeae and T. pallidum. If these STI microbial pathogens likewise make IgG crossreactive with T. vaginalis proteins A, E and G, and if a lysate of trichomonads is used for diagnosis, this may lead to the false identification of trichomonosis as the STI. Likewise, this work shows that a pathogen with immuno-crossreactive epitopes to human proteins is unsuitable as a vaccine. In this scenario, the immune response may lead to antibodies that contribute to autoimmune-like tissue cytopathology.
Future work should examine AEG::SOE3 as the target for IgG found in convalescent sera of patients who were infected with any of the other STI agents and other microbial pathogens mentioned above. A positive result would be a significant finding for several reasons. First, epitope mapping would show the epitopes of AEG::SOE3 detected by the sera of patients infected with other pathogens. Second, it would show that IgG to epitopes common to human and other microorganism proteins is more common than previously appreciated. Third, it may be possible to determine whether infections with certain microbial pathogens, such as those that cause STIs, make serum IgG to a common set of epitopes. This is because the nature of the extent of crossreactivity to AEG::SOE3 among infectious diseases may mean we must reconsider metabolic enzymes as virulence factors and possible mediators of disease pathogenesis.
Finally, this work highlights the possibility of constructing a novel SOE protein with epitopes found to be common among select groupings of microbial pathogens. For example, in the case of T. vaginalis, C. trachomatis, N. gonorrhoeae, and T. pallidum, a chimeric protein comprised of epitopes with 100% sequence identity could represent a broad-spectrum diagnostic that can be used in large at-risk populations. Positive results could then lead to additional specific diagnosis of the STI pathogen or multiple STI agents for treatment and cure. As it is recognized that multiple STIs can exist within patients [26], this type of diagnostic may have merit in screening of at-risk populations.
In the context mentioned above, we need a new paradigm when we find IgG to metabolic enzymes during immune responses to infectious diseases. When considering that trichomonosis is a significant asymptomatic disease and that women and men may be infected for long periods of time, it is reasonable to hypothesize that a positive serum IgG test to AEG::SOE3 indicates the presence of immuno-crossreactive antibodies, which may have consequences heretofore not considered for this STI in disease pathogenesis.
Conclusion
This paper presents a new epitope chain protein (AEG::SOE3) made up of immunogenic epitopes that are common and have sequence identity to humans and other bacteria and fungi, including three organisms that cause STIs. Data show that patients with trichomonosis make serum IgG antibody to A, E and G proteins. The work highlights the possibility of immuno-crossreactive antibody to human proteins, which may lead to disease pathogenesis. The work underscores that, in the future, an SOE protein can be constructed as a diagnostic that identifies multiple, related infectious agents, such as the four microorganisms causing STIs in this study, which then permits more specific diagnosis of single or multiple organisms causing disease.
Patents
• No. 9910042, “Strings of Epitopes Useful in Diagnosing
and Eliciting Immune Responses to Sexually Transmitted
Infections.” Inventor J.F. Alderete, Issued to Washington State
University March 5, 2018.
• No. 10386369, “Strings of Epitopes Useful in Diagnosing
and Eliciting Immune Responses to Sexually Transmitted
Infections.” Inventor J.F. Alderete, Issued to Washington State
University August 20, 2019.
Ethical approval for use of recombinant E. coli and experimental use of sera
Approval was obtained by the Washington State University Biosafety Approval of the Institutional Review Board (number 01058) for the use of sera, preparation of monoclonal antibodies using immunized mice, and use of recombinant E. coli
Acknowledgement
I thank a past collaborator at Washington University at St. Louis, MO for providing sera that was seropositive for α-actinin and that permitted us to screen for epitopes of the enzymes that form the basis of AEGSOE3. I also thank Ob/Gyn researchers at the University of Texas Health Science Center-San Antonio for providing women and men patient sera used in prior studies by my laboratory there and used here at
Washington State University. Ethical approval was given to use the sera by Institutional Review Boards at Washington U. as well as Washington State University. I want to acknowledge the help of Grace Alderete for maintenance of the laboratory during this research.
Conflict of Interest
The author declares no conflicts of interest. I alone designed the study and was responsible in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Hobbs MM, Sena AC, Swygard H, Schwebke JR (2008) Trichomonas vaginalis and trichomoniasis. Holmes KK, Sparling PF, Stamm WE, Piot P, Wasserheit JN, et al. Eds. Sexually Transmitted Diseases, McGraw-Hill Medical, New York, USA.
- Swygard H, Sena AC, Hobbs M, Cohen M (2004) Trichomoniasis: clinical manifestations, diagnosis, and management. Sex Transm Infect 80(2): 91-95.
- Gaydos CA, Klausner JD, Pai NP, Kelly H, Coltart C, et al. (2017) Rapid and point-of-care tests for the diagnosis of Trichomonas vaginalis in women and men. Sex Transm Infect 93(S4): S31-S35.
- Alderete JF (2020) Advancing Prevention of STIs by developing specific serodiagnostic targets: Trichomonas vaginalis as a model. Int J Environ Res Public Health 17(16): 5783.
- Neace CJ, Alderete JF (2013) Epitopes of the highly immunogenic Trichomonas vaginalis α-actinin are serodiagnostic targets for both women and men. J Clin Microbiol 51(8): 2483-2490.
- Martinez Galiano JM, Delgado Rodriguez M (2021) The relegated goal of health institutions: sexual and reproductive health. Int J Environ Res Public Health 18(4): 1767-1771.
- Alderete JF (2017) Epitopes within recombinant α-actinin protein is serodiagnostic target for Trichomonas vaginalis sexually transmitted infections. Heliyon 3(1): e00237.
- Alderete JF, Neace CJ (2013) Identification, characterization, and synthesis of peptide epitopes and a recombinant six-epitope protein for Trichomonas vaginalis serodiagnosis. Immunotargets Ther 2: 91-103.
- Bogdanos DP, Smyk DS, Invernizzi P, Rigopoulou EI, Blank M, et al. (2013) Infectome: a platform to trace infectious triggers of autoimmunity. Autoimmunity Review 12(7): 726-740.
- Floreani A, Restrepo Jimenez P, Secchi MF, De Martin S, Leung PSC, et al. (2018) Etiopathogenesis of autoimmune hepatitis. J Autoimmun 95: 133-143.
- Kirdar S, Sener AG, Cengiz M, Aydin N (2016) The prevalence of autoantibody and its relationship with genotypes of hepatitis C virus in patients with chronic hepatitis C virus infection. APMIS 124(11): 979-984.
- Amedei A, Bergman MP, Appelmelk BJ, Azzurri A, Benagiano M, et al. (2003) Molecular mimicry between Helicobacter pylori antigens and H+,K+-adenosine triphosphate inhuman gastric autoimmunity. J Expt Med 198(8): 1147-1156.
- Cunha Neto E, Coelho V, Guilherme L, Fiorelli A, Stolf N, et al. (1996) Autoimmunity in Chagas’ disease. Identification of cardiac myosin-B13 Trypanosoma cruzi protein crossreactive T cell clones in heart lesions of chronic Chagas’ cardiomyopathy patient. J Clin Invest 98(8): 1709-1712.
- Da Rocha Sobrinho JM, Jarach R, da Silva NA, Shio MT, Jancar S, et al. (2011) Mycoplasmal lipidassociated membrane proteins and Mycoplasma arthritidis mitogen recognition by serum antibodies from patients with rheumatoid arthritis. Rheumatol Int 31(7): 951-957.
- Abdulkarim AS, Petrovic LM, Kim WR, Angulo P, Lloyd RV, et al. (2004) Primary biliary cirrhosis: an infectious disease caused by Chlamydia pneumoniae? J Hepatol 40(3): 380-384.
- Liu HY, Deng AM, Zhang J, Zhou Y, Yao DK, et al. (2005) Correlation of Chlamydia pneumoniae infection with primary biliary cirrhosis. World J Gastroenterol 11(26): 4108-4110.
- Christen U, Hintermann E (2014) Pathogen infection as a possible cause for autoimmune hepatitis. Int Rev Immunol 33(4): 296-313.
- Erickson TA, Muscal E, Munoz FM, Lotze T, Hasbun R, et al. (2020) Infectious and autoimmune causes of encephalitis in children. Pediatrics 145(6): 1-8.
- Alderete JF (1983) Antigenic analysis of several pathogenic strains of Trichomonas vaginalis. Infect Immun 39(3): 1041-1047.
- Mundodi V, Kucknoor AS, Alderete JF (2008) α-Enolase is a surface-associated plasminogen-binding protein of Trichomonas vaginalis. Infect Immun 76(2): 523-531.
- Lama A, Kucknoor A, Mundodi V, Alderete JF (2009) Glyceraldehyde-3-phosphate dehydrogenase is a surface-associated, fibronectin-binding protein of Trichomonas vaginalis. Infect Immun 77(7): 2703-2711.
- Sunblad V, Bussmann L, Chiauzz, VA, Pancholi V, Charreau EH (2006) Alpha-enolase: a novel autoantigen in patients with premature ovarian failure. Clin Endocrinol 65(6): 745-751.
- Kim SR, Kim JH, Park SJ, Lee HY, Kim YS, et al. (2015) Comparison between mixed lysate antigen and α-actinin antigen in ELISA for serodiagnosis of trichomoniasis. Parasitol Int 64(5): 405-407.
- Cudmore SL, Garber GE (2010) Prevention or treatment: the benefits of Trichomonas vaginalis J Infect Public Health 3(2): 47-53.
- Smith, J, Garber GE (2014) Current status and prospects for development of a vaccine against Trichomonas vaginalis Vaccine 32(14): 1588-1594.
- Pakianathan MR, Ross JDC, McMillan A (1996) Characterizing patients with multiple sexually acquired infections: a multivariate analysis. Int J STD & AIDS 7(5): 359-360.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.