Mini Review 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Removing Dental Plaque Biofilm: A Review of Dextranases
*Corresponding author: Lyu M and Shujun Wang, Jiangsu Key Laboratory of Marine Bioresources and Environment, Jiangsu Ocean University, China.
Received: June 07, 2021; Published: June 23, 2021
DOI: 10.34297/AJBSR.2021.13.001862
Abstract
Various types of resident microorganisms are colonized in the human oral cavity, and they are mainly probiotics that are beneficial to the health of the host. These bacteria and foreign temporary bacteria mainly gather in the dental plaque biofilm on the tooth surface. Dextranase is a glycosidic bond hydrolase, which specifically hydrolyzes the α-1,6 glycosidic bond of dextran that is the mainly component of the biofilm and can be used to remove dental plaque biofilm. This article mainly reviews the removal of dental plaque biofilm by dextranase.
Keywords: Dextranase; Dental Plaque; Biofilm; Streptococcus Mutans
Introduction
The human oral cavity is a very suitable environment for microbial colonization. Studies have found that these microorganisms mainly accumulate on the biofilm (plaque) on the tooth surface. Dental plaque biofilm is a chronic bacterial disease that occurs on or inside teeth. It is characterized by high incidence and wide distribution, which seriously endangers human health, especially oral health. Removal of these plaques can effectively reduce the adhesion of microorganisms to the surface of the plaque mucosa. Streptococcus mutans presented in the human oral cavity ferments the residual sucrose in the oral cavity to produce water-soluble dextran with α-1,6, α-1,3 glucan bond as the main extracellular polysaccharide. Controlling dextran to reduce the formation of dental plaque biofilm has become an important means to prevent dental caries. This review mainly focuses on the removal of dental plaque biofilm with dextranase.
The Formation of Dental Plaque Biofilm
There are a large number of microorganisms in the oral cavity of animals [1]. Dental plaque biofilm is composed of many microbial flora and adheres to the surface of teeth, which has an important impact on oral health. many oral diseases are related to the formation of dental plaque [2,3]. At present, S. mutans plays significant role in the forming of dental caries. Therefore, S. mutans is often used as a model strain in researching and constructing dental plaque biofilms [4]. The formation of dental plaque biofilm changes dynamically with time. Kolenbrander found that the initial stage of dental plaque biofilm is mainly Streptococcus, and the later stage is mainly Bacillus, Filamentous and Actinomycetes [5]. S. mutans, S. sobrinus and S. sanguis are the early strains of dental plaque biofilm formation and play a key role in the formation of dental caries [4,6]. Alahmad found that the main component of dental plaque biofilm was Streptococcus on the first day [7]. In addition, studies have found that dead bacteria are also one of the components in the biofilm.
In the early process of dental plaque biofilm formation, they compete with live bacteria for contact positions, thereby promoting the formation of biofilm [8]. In the early days, the adsorption of biofilm was implemented by Streptococcus mainly, which produced adhesin to bind to the protein receptors on the formed tooth-acquired membrane and colonized the tooth surface [9]. Streptococcus can produce three kinds of glucosyltransferases, namely GtfD that synthesizes water-soluble α-1,6-glucan, GtfB that synthesizes water-insoluble α-1,3-glucan, and a mixture of two glucans. GtfC, they can quickly synthesize extracellular polysaccharides from dietary sucrose [9-11]. These two extracellular polysaccharides are connected in an orderly spiral form to form a dental plaque matrix, provided a colonization site for other oral microorganisms, thereby other oral microorganisms were promoted to colonize the dental plaque matrix. Finally, a plaque biofilm was formed and attached firmly on a surface of the tooth [12,13].
The Source and Characteristics of Dextranase
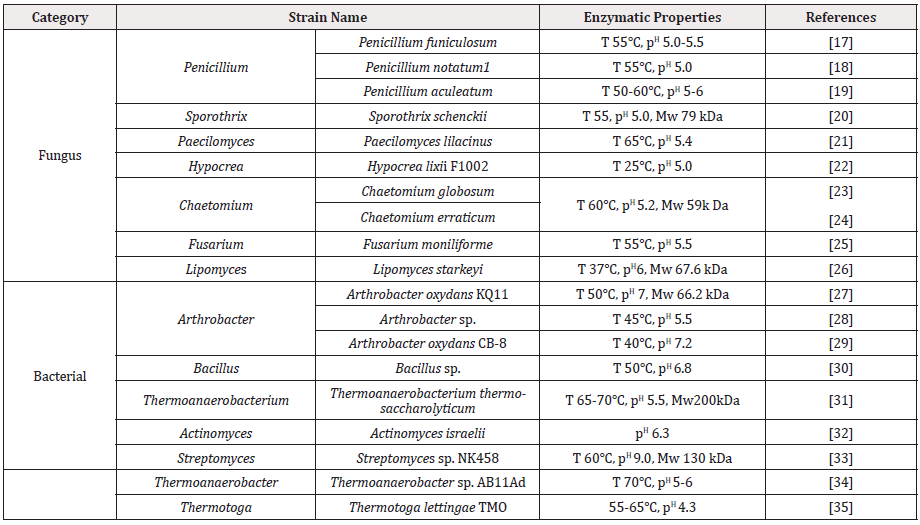
Dextranase has been found in animals, plants, and microorganisms. The most reported dextranases are from microorganisms, including fungi and bacteria. Compared with dextranase derived from yeast and bacteria, molds usually have higher enzyme activity [14]. The dextranases produced by Chaetomium and Penicillium have been used in industrial production. P. lilacinum NRRL896 and P. funiculosum NRRL1132 have been reported they could produce dextranases that have relatively high optimal reaction temperature. Furthermore, the optimal pH of reaction was in slightly acidic condition. The characteristics of dextranase from molds were listed in Table 1&[15]. There are different sources of bacteria that could produce dextranase, and the enzymatic properties are also quite different. The time of bacterial fermentation to produce dextranase is shorter than that of mold fermentation, generally about 2 days. With less secondary metabolites, it would be conducive to the purification of dextranase in the later stage, and has little toxicity [16]. The properties of dextranase secreted form bacteria was listed in Table 1 [17-25]. The optimal of temperature and pH were diversity.
Dextranase Removing Dental Plaque
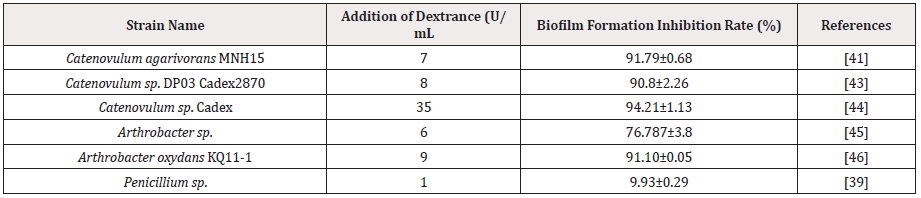
Dextranase has represented excellent effect value in the prevention and removal of dental plaque biofilm [26-35]&[Table 2]. As early as 1971, Caldwell and Robert conducted related studies on adding dextranase to mouthwashes to treat dental plaque biofilm [36]. In 1972, Keyes reported that dextranase can effectively remove dental plaque, and researchers continue to study the inhibitory effect of dextranase on dental plaque [37]. In 2002, Marotta hydrolyzed the water-insoluble glucan produced by S. sobrinus with commercial dextranase and also eliminated glucan that had already formed [38]. In recent years, more and more researchers have carried out research on dextranase to remove and defend dental plaque biofilm. Qiu used S. mutans, Lactobacillus acidophilus and Actinomycetes to establish a biofilm system in vitro, and then he added dextranase, sodium fluoride and the mixture to treat the biofilm. It was found that the biofilm treated with the mixture was very thoroughly damage [39,40] discovered the dextran-dependent aggregation of S. mutans through microarray analysis of clinical strains. When dextranase was added, S. mutans also cannot aggregate, thereby the formation of dental plaque biofilm was reduced significantly [40].
Xiaohua Lai isolated the marine bacterium Catenovulum agarivorans MNH15 that could secret dextranase. The study showed that the dextranase could be very effective inhibited the formation of dental plaque biofilm. Furthermore, there was a good removal effect on the dental plaque biofilm that had already shaped by S. mutans [41]. Otsuka linked the mutanase gene derived from Paenibacillus humicus NA1123 and the dextranase gene derived from S. mutans ATCC 25175 into the vector at the same time and the E. coli expressed a chimera with two enzyme functions. The chimeric enzyme could degrade water-insoluble glucan and it expressed efficiency ability to destroy the biofilm. Moreover, removal rate was over 4 times higher than the mixture of dextranase and mutanase [42]. At present, most of the oral care products on the market are added with antibacterial agents such as alcohol, lysozyme, while products containing non-antibacterial dextranase are rare [43-46]. Some companies in the United States and Japan are mainly producing oral care products containing dextranase. Compared with the mechanical method and the chemical drug method, the biological enzyme method has significant advantages such as safety, effectiveness and low price. Therefore, the research and development of adding biological enzymes in oral care products have broad prospects.
Conclusion
The unlimited construction of dental plaque biofilm is a natural phenomenon that exists in everyone's mouth. As a result, S. mutans causes severe dental caries, dental plaque and endocarditis, which seriously affects human health. Therefore, necessary precautions need to be taken to avoid these troublesome infections. For oral care products, they must have a tooth-protecting effect without disturbing the natural environment of probiotics in mouth. The inhibitory effect of dextranase on S. mutans has been confirmed. The dextranase that is appropriately added in oral care products research should be selective and targeted. The optimal temperature and pH of dextranase are important, and the dextranase will not be affected by other additives of oral care products. Furthermore, the stability of dextranase in room temperature need to focus on. Dextranases have broad prospective to protect human oral health of the microenvironment of human mouth.
Conflict of Interest
The authors confirm that there is no conflict of interests regarding this paper.
Acknowledgements
This study was supported by the National Key R&D Program of China (2018YFC0311106). The Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
- Gurenlian JAR (2007) The Role of Dental Plaque Biofilm in Oral Health. Journal of dental hygiene. American Dental Hygienist’s Association 81(5): 116-116.
- Marsh PD (2010) Microbiology of Dental Plaque Biofilms and Their Role in Oral Health and Caries. Dental Clinics of North America 54(3): 441-454.
- Jenkinson HF, RJ Lamont (2005) Oral microbial communities in sickness and in health. Trends in Microbiology 13(12): 589-595.
- Beighton D (2010) The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dentistry & Oral Epidemiology 33(4): 248-255.
- Kolenbrander PE, Roxanna N Andersen, David S Blehert, Paul G Egland, Jamie S Foster, et al. (2002) Communication among oral bacteria. Microbiol Mol Biol Rev 66(3): 486-505.
- Salam MA, Naoko Matsumoto, Khairul Matin, Yuzo Tsuha, Ryoma Nakao, et al. (2004) Establishment of an Animal Model Using Recombinant NOD.B10.D2 Mice To Study Initial Adhesion of Oral Streptococci. Clinical Diagnostic Laboratory Immunology 11(2): 379-386.
- Al-Ahmad A, Axel Wunder, Thorsten Mathias Auschill, Marie Follo, Gabriele Braun, et al. (2007) The in vivo dynamics of Streptococcus spp., Actinomyces naeslundii, Fusobacterium nucleatum and Veillonella spp. in dental plaque biofilm as analysed by five-colour multiplex fluorescence in situ hybridization. Journal of Medical Microbiology 56(5): 681-687.
- Hope CK, M Wilson (2006) Biofilm structure and cell vitality in a laboratory model of subgingival plaque. Journal of Microbiological Methods 66(3): 390-398.
- Carlén A, J Olsson, P Ramberg (1996) Saliva mediated adherence, aggregation and prevalence in dental plaque of Streptococcus mutans, Streptococcus sanguis and Actinomyces spp, in young and elderly humans. Archives of Oral Biology 41(12): 1133-1140.
- Hanada N, HK Kuramitsu (1989) Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infection & Immunity 57(7): 2079-2085.
- H Aoki, T Shiroza, M Hayakawa, S Sato, HK Kuramitsu (1986) Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infection & Immunity 53(3): 587-594.
- William H Bowen, Robert A Burne, Hui Wu, Hyun Koo (2018) Oral Biofilms: Pathogens, Matrix and Polymicrobial Interactions in Microenvironments. Trends in Microbiology 26(3): 229-242.
- Tanya L Johnson, Jiunn C Fong, Chelsea Rule, Andrew Rogers, Fitnat H Yildiz, et al. (2014) The Type II Secretion System Delivers Matrix Proteins for Biofilm Formation by Vibrio cholerae. Journal of Bacteriology 196(24): 4245-4252.
- Ding Tao, WuHong Bin Zhang, Li Jun Huang, Xue Qin Hu (2011) Purification and characterization of extracellular dextranase from a novel producer, Hypocrea lixii F1002 and its use in oligodextran production. Process Biochemistry 46(10): 1942-1950.
- Hutson DH, H Weigel (1963) Studies on dextrans and dextranases 4. Mechanism of the actions of intra- and extra-cellular mould hydrolases. Biochemical Journal 88(3): 588-591.
- Cai R, Mingsheng Lu, Yaowei Fang, Yuliang Jiao, Qiang Zhu, et al. (2014) Screening, production, and characterization of dextranase from Catenovulum sp. Annals of Microbiology. 64(1): 147-155.
- N Kosaric, K Yu, J E Zajic (2010) Dextranase production from Penicillium funiculosum Biotechnol Bioeng 15(4): 729-741.
- Szczodrak J, M Pleszczyńska, J Fiedurek (1994) Penicillium notatum1 a new source of dextranase. Journal of Industrial Microbiology 13(5): 315-320.
- Madhu, KA Prabhu (1984) Studies on dextranase from Penicillium aculeatum Enzyme and Microbial Technology 6(5): 217-220.
- Arnold WN, TBP Nguyen, LC Mann (1998) Purification and characterization of a dextranase from Sporothrix schenckii Archives of Microbiology 170(2): 91-98.
- Galvez Mariscal A, Lopez Munguia (1991) Production and characterization of a dextranase from an isolated Paecilomyces lilacinus strain 36(3): 327-331.
- Ding-Tao Wu, Hong-Bin Zhang, Li-Jun Huang, Xue Qin (2011) Hu Purification and characterization of extracellular dextranase from a novel producer, Hypocrea lixii F1002, and its use in oligodextran production. Process Biochemistry 46(10): 1942-1950.
- Yang, L, N Zhou, Y Tian (2019) Characterization and application of dextranase produced by Chaetomium globosum mutant through combined application of atmospheric and room temperature plasma and ethyl methyl sulfone. Process Biochemistry 85: 116-124.
- JJ Virgen-Ortíza, V Ibarra-Junqueraa, P Escalante-Minakataa, JA Osuna-Castroc, A González-Potesa (2015) Kinetics and thermodynamic of the purified dextranase from Chaetomium erraticum. Journal of Molecular Catalysis B: Enzymatic 122: 80-86.
- Simonson LG, AE Liberta, A Richardson (1975) Characterization of an Extracellular Dextranase from Fusarium moniliforme. Applied Microbiology 30(5): 855-861.
- Hee-Kyoung Kang, Seung Heuk Kim, Ji-Young Park, Xing-Ji Jin, Deok-Kun Oh, et al. (2005) Cloning and characterization of a dextranase gene from Lipomyces starkeyi and its expression in Saccharomyces cerevisiae. Yeast 22(15): 1239-1248.
- Wang D, Mingsheng Lu, Shujun Wang, Yuliang Jiao, Weijuan L, et al. (2014) Purification and characterization of a novel marine Arthrobacter oxydans KQ11 dextranase. Carbohydrate Polymers 106: 71-76.
- Yu-Liang Jiao, Shu-Jun Wang, Ming-Sheng Lv, Bing-Hua Jiao, Wei-Juan Li, et al. (2014) Characterization of a marine-derived dextranase and its application to the prevention of dental caries. Journal of Industrial Microbiology & Biotechnology 41(1): 17-26.
- Jin HL, Seong Hee Nam, Hyen Joung Park, Young-Min Kim, Nahyun Kim, et al. (2010) Biochemical Characterization of Dextranase from Arthrobacter oxydans and Its Cloning and Expression in Escherichia coli. Food Science & Biotechnology 19(3): 757-762.
- Elvira Khalikova, Petri Susi, Nikolai Usanov, Timo Korpela (2003) Purification and properties of extracellular dextranase from a Bacillus sp. Journal of Chromatography B 796(2): 315-326.
- Frank H, D Rolf, G Gerhard (2001) Isolation of a new Thermoanaerobacterium thermosaccharolyticum strain (FH1) producing a thermostable dextranase. Journal of General & Applied Microbiology 47(4): 187-192.
- Staat RH, CF Schachtele (1975) Characterization of a dextranase produced by an oral strain of Actinomyces israelii. Infection & Immunity 12(3): p. 556-563.
- Shweta Purushe, Divya Prakash, Neelu N Nawani, Prashant Dhakephalkar, Balasaheb Kapadnis, et al. (2012) Biocatalytic potential of an alkalophilic and thermophilic dextranase as a remedial measure for dextran removal during sugar manufacture 115: 2-7.
- Wynter C, BK Patel, P Bain, J de Jersey, S Hamilton, et al. (1996) A novel thermostable dextranase from a Thermoanaerobacter species cultured from the geothermal waters of the Great Artesian Basin of Australia. Fems Microbiology Letters 40(2): 271-276.
- Kim YM, D Kim (2010) Characterization of novel thermostable dextranase from Thermotoga lettingae TMO 85(3): 581-587.
- Caldwell RC, HJ Sandham, WV Mann, SB Finn, AJ Formicola, et al. (1971) The effect of a dextranase mouthwash on dental plaque in young adults and children. Journal of the American Dental Association 82(1): 124-131.
- PH Keyes, MA Hicks, M Goldman, RM McCabe, RJ Fitzgerald (1971) Dispersion of dextranous bacterial plaques on human teeth with dextranase. Journal of the American Dental Association 82(1): 136-141.
- M Pleszczyńska, A Wiater, J Szczodrak (2002) Degradation of dental plaque glucans and prevention of glucan formation using commercial enzymes. Process Biochemistry 38(1): 101-108.
- Yuan Xin Qiu, Meng-Ying Mao, Dan Jiang, Xiao Hong, Ying-Ming Yang (2016) Co-operative effect of exogenous dextranase and sodium fluoride on multispecies biofilms. Journal of Dental ences 11(1): 41-47.
- Senpuku H, Hideo Yonezawa, Saori Yoneda, Itaru Suzuki, Ryo Nagasawa, et al. (2017) SMU.940 regulates dextran‐dependent aggregation and biofilm formation in Streptococcus mutans. Molecular Oral Microbiology 33(1): 47-58.
- Xiaohua Lai, Xin Liu, Xueqin Liu, Tian Deng, Yanli Feng (2019) The Marine Catenovulum agarivorans MNH15 and Dextranase: Removing Dental Plaque. Marine Drugs 17(10): 592.
- Otsuka R, Susumu Imai, Takatoshi Murata, Yoshiaki Nomura, Masaaki Okamoto, et al. (2015) Application of chimeric glucanase comprising mutanase and dextranase for prevention of dental biofilm formation. Microbiology & Immunology 59(1): 28-36.
- Tian Deng, Yanli Feng, Linxiang Xu, Xiaopeng Tian, Xiaohua Lai, et al. (2020) Expression, purification and characterization of a cold-adapted dextranase from marine bacteria and its ability to remove dental plaque. Protn Expression and Purification 174: 105678.
- Wei Ren, Ruanhong Cai, Wanli Yan, Mingsheng Lyu, Yaowei Fang, et al. (2018) Purification and Characterization of a Biofilm-Degradable Dextranase from a Marine Bacterium. Marine Drugs 16(2): 51.
- Yu Liang Jiao, Shu Jun Wang, Ming Sheng Lv, Bing Hua Jiao, Wei Juan Li, et al. (2014) Characterization of a marine-derived dextranase and its application to the prevention of dental caries. Journal of Industrial Microbiology & Biotechnology 41(1): 17-26.
- Xiaobei Wang, Huaixu Cheng, Mingsheng Lu, Yaowei Fang, Yuliang Jiao, et al. (2016) Dextranase from Arthrobacter oxydans KQ11-1 inhibits biofilm formation by polysaccharide hydrolysis. Biofouling 32(9): 1223-1233.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.