Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
The Roles of BTG1 mRNA Expression in Cancers: A Bioinformatics Analysis
*Corresponding author: Hua Chuan Zheng, Department of Oncology and Experimental Center, The Affiliated Hospital of Chengde Medical University, Chengde, Hebei Province, China
Received: May 31, 2021; Published: June 10, 2021
DOI: 10.34297/AJBSR.2021.13.001840
Abstract
Background: BTG1 (B-cell translocation gene 1) might suppress proliferation and cell cycle progression, and induce differentiation, apoptosis, and anti-inflammation. This study aimed to clarify the clinicopathological and prognostic significances of BTG1 mRNA expression in cancers.
Methods: We performed a bioinformatics analysis of BTG1 mRNA expression through Oncomine, TCGA and Kaplan-Meier plotter databases up to July 1, 2017.
Results: Oncomine data showed that BTG1 expression was lower in gastric cancer than normal mucosa, but versa for pulmonary squamous cell carcinoma, breast invasive ductal cancer and ovarian cancer (p<0.05). In term of TCGA, BTG1 expression was positively correlated with dedifferentiation, histological grading, and poor prognosis of gastric cancer (p<0.05) and TNM staging of ovarian cancer (p<0.05), but negatively associated with lymph node metastasis, TNM staging and adverse prognosis of breast cancer (p<0.05). BTG1 expression was higher in pulmonary squamous cell carcinoma than adenocarcinoma (p<0.05). Younger age, lymph node metastasis, TNM staging and BTG1 hypoexpression was independent factors for worse prognosis of the breast cancer patients (p<0.05). According to Kaplan-Meier plotter, BTG1 expression was negatively correlated with favorable prognosis of the gastric, lung or ovarian cancer patients, but the converse was true for breast cancer patients.
Conclusion: BTG1 expression might be employed as a potential marker to indicate carcinogenesis and subsequent progression, even prognosis.
Keywords: BTG1, Bioinformatics analysis, Carcinogenesis, Aggressiveness, Prognosis
Introduction
BTG1 (B-cell translocation gene 1) is reported to suppress cell proliferation and cell cycle progression and induce cell differentiation due to its interaction with the myogenic factor MyoD [1], protein arginine methyltransferase 1 [2], and human carbon catabolite repressor protein-associative factor 1 [3]. BTG1 has also been reported to enhance Hoxb9-induced transcription to suppress proliferation in HeLa cells [4]. Additionally, BTG1 mediates apoptotic induction, which is evidenced by BTG1 overexpression in apoptotic cells [5] and the contribution of BTG1 to anti-sense Bcl-2- induced cytotoxic effects [6]. Liu et al. [7] found that BTG1 potentiated apoptosis and suppressed proliferation in renal cell carcinoma by interacting with PRMT1. BTG1 could reverse the miR-22-induced inhibition of autophagy [8] and miR-4295 significantly promoted proliferation, colony formation, and migration of bladder cancer cell via directly targeting BTG1 [9]. MiRNA-511 promotes the proliferation of human hepatoma cells and miRNA 301A promotes colitis-associated cancer development by inhibiting BTG1 [10,11]. BTG1 functioned as a direct target of miR-330-3p, and miR-27a-3p in hepatocellular carcinoma and ovarian cancer cells, and thereby weakened cell viability, migration, and invasion, and promoted cell apoptosis [12,13]. BTG1 was shown to prevented antigen from inducing molecular features of in vitro allergic reactions as a direct target of miR-183-5p. In atopic dermatitis, NF-κB overexpression and activation was shown to promote the transcription of miR-183-5p by binding to its promoter [14].
Su et al. [15] reported that BTG1 overexpression triggered G1/S phase cell cycle arrest and increased apoptosis in HCT-116 cells via the ERK/MEK signaling pathway. Zhu et al. [16] reported that BTG1 enhanced the radiation sensitivity of human breast cancer by inducing cell cycle arrest, the formation of reactive oxygen species, chromosomal aberrations, and apoptosis via inhibition of the PI3K/Akt signaling pathway. BTG1 overexpression was also found to suppress proliferation, tumor growth and lung metastasis, induce differentiation, autophagy, and apoptosis, and mediate chemosensitivity in colorectal and gastric cancer cells [17,18]. In combination of these data, it is suggested that BTG1 may function as a tumor suppressor. In the present study, we aimed to clarify the clinicopathological and prognostic significances of BTG1 mRNA expression in cancers by a bioinformatics analysis of high-throughput cDNA array and RNA sequencing uding online Oncomine, TCGA and Kaplan-Meier plotter.
Materials and Methods
Oncomine Database Analysis
The individual gene expression level of BTG1 mRNA was analyzed using Oncomine (www.oncomine.org), a cancer microarray database and web-based data mining platform for a new discovery from genome-wide expression analyses. We compared the differences in BTG1 mRNA level between normal tissue and cancer. All data were log-transformed, median centered per array, and standard deviation normalized to one per array.
TCGA Database Analysis
The expression data (RNA-seqV2) and clinicopathological data of gastric (n=392), lung (n=865), breast (n=1093) and ovarian (n=304) cancer patients were downloaded from the Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) database by TCGA-assembler in R software [19]. We integrated the raw data, analyzed BTG1 expression in the cancers, and compared it with clinicopathological and prognostic data of the cancer patients. The means were compared with student t test. Kaplan-Meier survival plots were generated with survival curves compared by log-rank statistic. Cox’s proportional hazards model was employed for multivariate analysis. Two-sided p<0.05 was considered as statistically significant. SPSS 17.0 software was employed to analyze all data.
Kaplan-Meier Plotter Analysis
The prognostic significance of BTG1 mRNA was also analyzed in gastric, lung, breast and ovarian cancers using Kaplan-Meier plotter (http://kmplot.com) [20].
Results and Discussion
The Clinicopathological and Prognostic Significances of BTG1 mRNA Expression in Gastric Cancer
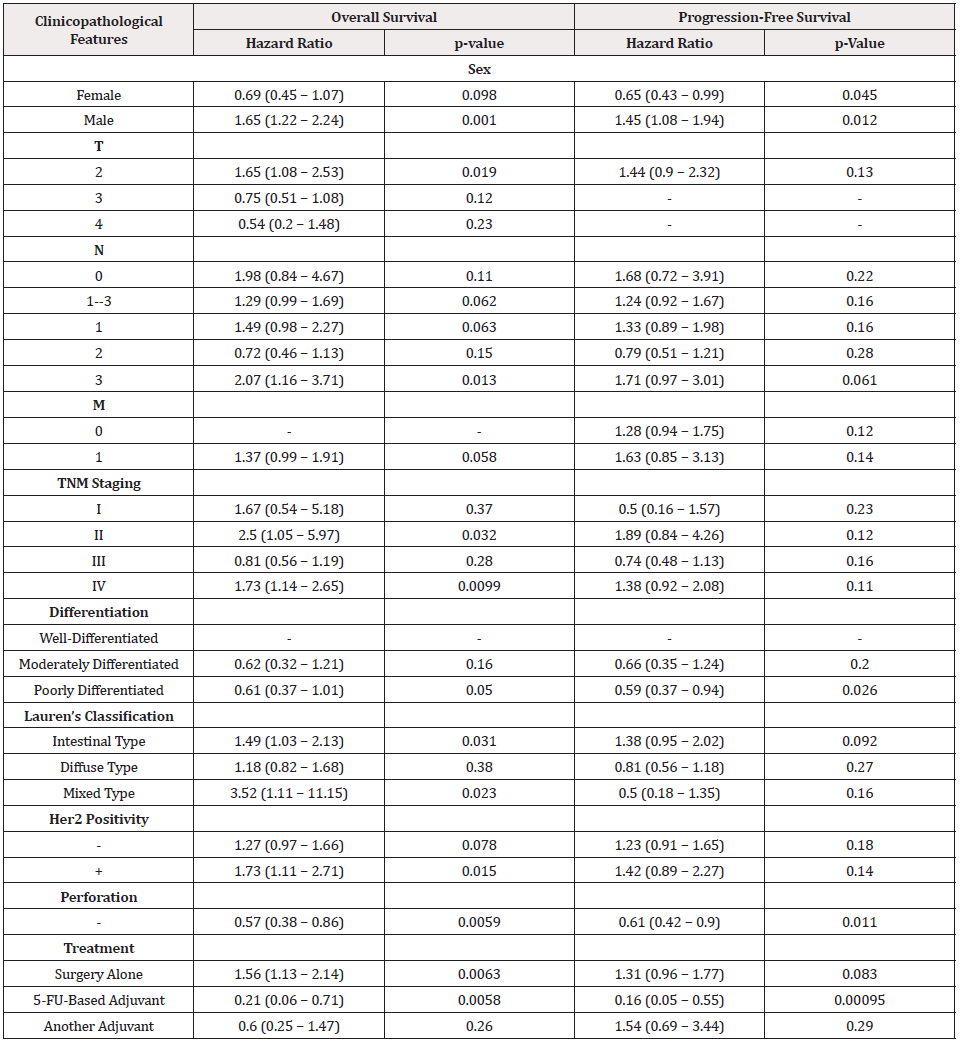
According to Wang’s database, we found that BTG1 mRNA expression was lower in gastric cancer than normal tissues (Figure 1A, p<0.05). In TCGA data, BTG1 expression was positively correlated with dedifferentiation, histological grading, and poor prognosis of gastric cancer (Figure 1B, p<0.05). According to Kaplan-Meier plotter, a higher BTG1 expression was negatively correlated with overall and progression-free survival rates of all cancer patients, male or perforating cancer patients and the patients receiving 5-FU-based adjuvant (Figure 1C, Figure 1D &Table 1, p<0.05). As shown in Table 1, stage II and IV, T2, N3, intestinal- and mixed-type, or Her2-positive cancer patients with high BTG1 expression showed a shorter overall survival time than those with its low expression (p<0.05). It was similar for progression-free survival in female, male, or poorly differentiated cancer patients (p<0.05). Negative association between BTG1 expression and overall prognosis was observed in the cancer patients only receiving surgical operation (Table 1, p<0.05).
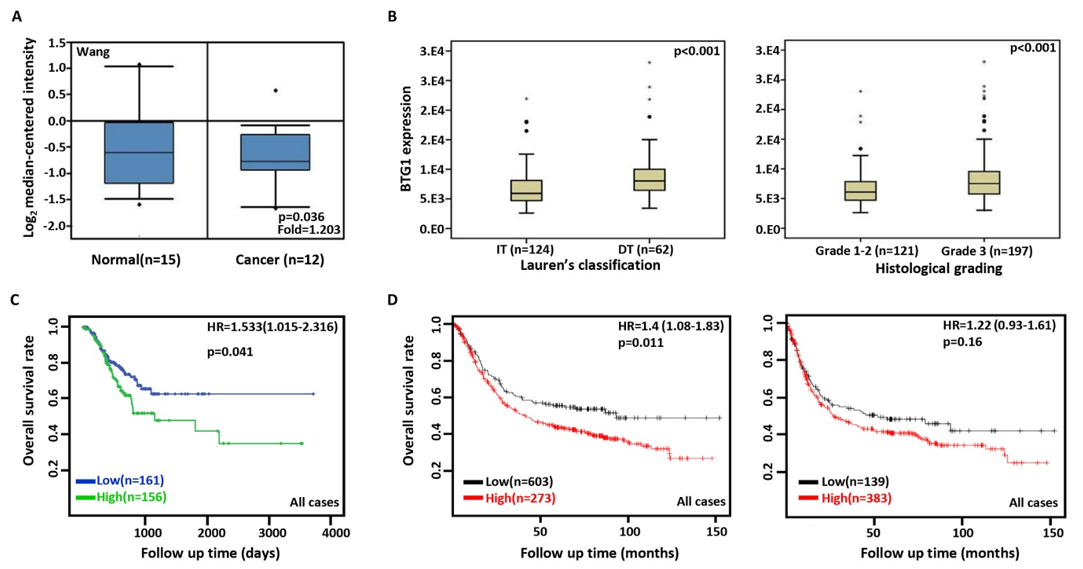
Figure 1: The clinicopathological and prognostic significances of BTG1 mRNA expression in gastric cancer.
Wang’s dataset was used for bioinformatics analysis to explore BTG1 expression in gastric cancer. A lower BTG1 expression was detectable in
gastric cancer than that in normal mucosa (A, p<0.05). TCGA database showed that BTG1 was less expressed in intestinal-type (IT) than diffusetype
(DT) ones, and in Grade 1-2 than Grade 3 carcinomas (B, p<0.05). BTG1 expression was negatively correlated with overall survival rate of the
cancer patients (C, p<0.05). It was the same for the overall and progression-free survival rates of the patients with gastric cancer according to the
data from Kaplan-Meier plotter (D, p<0.05). HR, hazard ratio.
The Clinicopathological and Prognostic Significances of BTG1 mRNA Expression in Lung Cancer
Then, we used Talbot's and Hou's datasets to perform bioinformatics analysis and found that BTG1 expression was higher in squamous cell carcinoma than normal tissue (Figure 2A, p<0.05). In TCGA data, BTG1 expression was higher in squamous cell carcinoma than adenocarcinoma, in male than female cancer patients, and in elder than younger cancer patients (Figure 2B, p<0.05). According to Kaplan-Meier plotter, we found that a higher BTG1 expression was negatively correlated with the overall rate of all, male, or grade 2 cancer patients (Figure 2C, p<0.05).
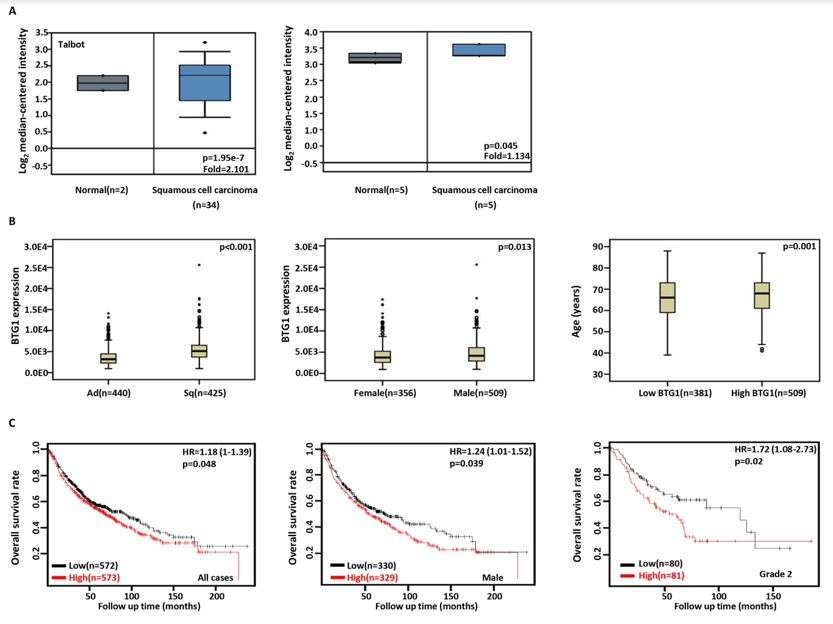
Figure 2: The clinicopathological and prognostic significances of BTG1 mRNA expression in lung cancer.
Talbot’s and Hou’s datasets were employed for bioinformatics analysis to analyze BTG1 expression during lung carcinogenesis. BTG1 expression
was up regulated in squamous cell carcinoma, compared with normal tissue (A, p<0.05). In TCGA database, BTG1 expression was compared
with histological subtyping, gender, and age of the cancer patients (B). The correlation between BTG1 expression and overall or post-progression
survival rate of the patients with lung cancer was analysis using KM plotter (C). HR, hazard ratio.
The Clinicopathological and Prognostic Significances of BTG1 mRNA Expression in Breast Cancer
BTG1 was more expressed in breast invasive ductal cancer than normal tissues according to Karnoub’s database (Figure 3A, p<0.05). TCGA database showed that BTG1 expression was negatively associated with lymph node metastasis, TNM staging and adverse prognosis of breast cancer (Figure 3B&3C, p<0.05). Cox’s risk proportional analysis showed that younger age, lymph node metastasis, TNM staging and BTG1 hypoexpression were independent factors for worse prognosis of the patients with breast cancer (Table 2, p<0.05). According to Kaplan-Meier plotter, a higher BTG1 expression was positively correlated with overall survival rates of all or luminal-B cancer patients (Figure 3D, p<0.05). Her2-negative and luminal-B cancer patients with high BTG1 expression showed a long distant- metastasis-free survival time than those with its low expression (p<0.05, data not shown). There appeared a positive relationship between BTG1 expression and the progression-free survival rate of the cancer patients without chemotherapy or margin invasion (p<0.05, data not shown). The overall survival rate of the patient with ER-negative, Grade-3, or luminal-B cancer was higher in the group of high BTG1 expression than its low expression (p<0.05, data not shown).
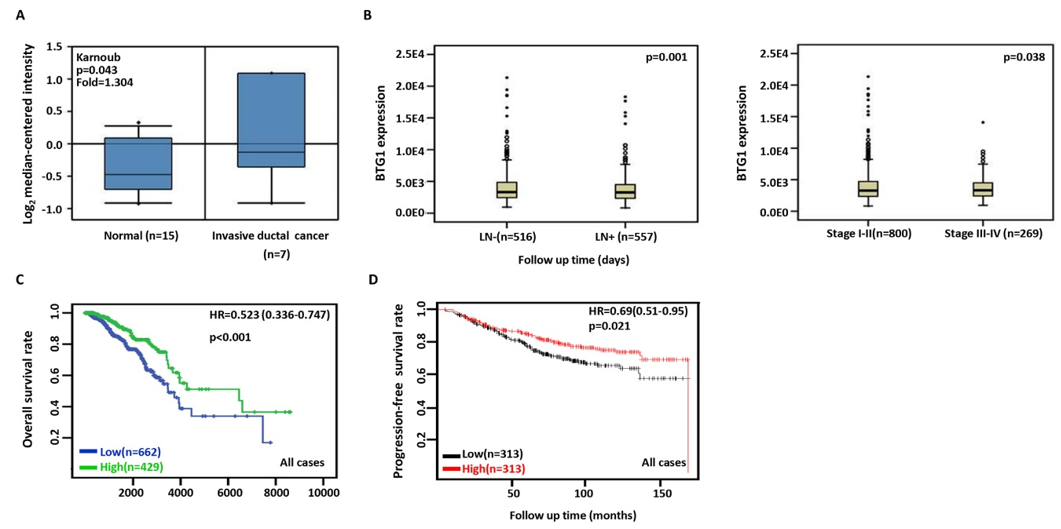
Figure 3: The clinicopathological and prognostic significances of BTG1 mRNA expression in breast cancer.
According to Karnoub’s data, BTG1 overexpression was detectable in breast invasive ductal cancer, compared with normal breast tissue (A, p<0.05). TCGA database showed a higher BTG1 expression was negatively correlated with lymph node metastasis and TNM staging of breast cancer (B, p<0.05). The overall survival rate of the cancer patients was higher in high than low BTG1 expression groups according to TCGA database (C, p<0.001) and KM plotter (D, p<0.05). HR, hazard ratio.

Table 2: Multivariate analysis of hazard factors of the prognosis of the patients with breast cancer.
Note*: Ad, Adenocarcinoma; Sq, Squamous Cell Carcinoma.
The Clinicopathological and Prognostic Significances of BTG1 mRNA Expression in Ovarian Cancer
Then, we used Bonome's and Hendrix’s datasets to perform bioinformatics analysis and found that BTG1 expression was higher in ovarian cancers or mucinous adenocarcinoma than normal tissues (Figure 4A, p<0.05). In TCGA data, BTG1 expression was higher in stage III-IV than I-II cancers (Figure 4B, p<0.05). According to Kaplan-Meier plotter, a higher BTG1 expression was negatively correlated with the overall survival rates of Dubulk suboptimal and p53-mutanttant cancer patients (Figure 4C, p<0.05). Stage I+II, II, II+III, III, and III+IV cancer patients with high BTG1 expression showed a short progression-free survival time than those with its low expression (Figure 4C, p<0.05). There appeared a negative relationship between BTG1 expression and the overall survival rate of the cancer patients with paclitaxel treatment (Figure 4D, p<0.05).
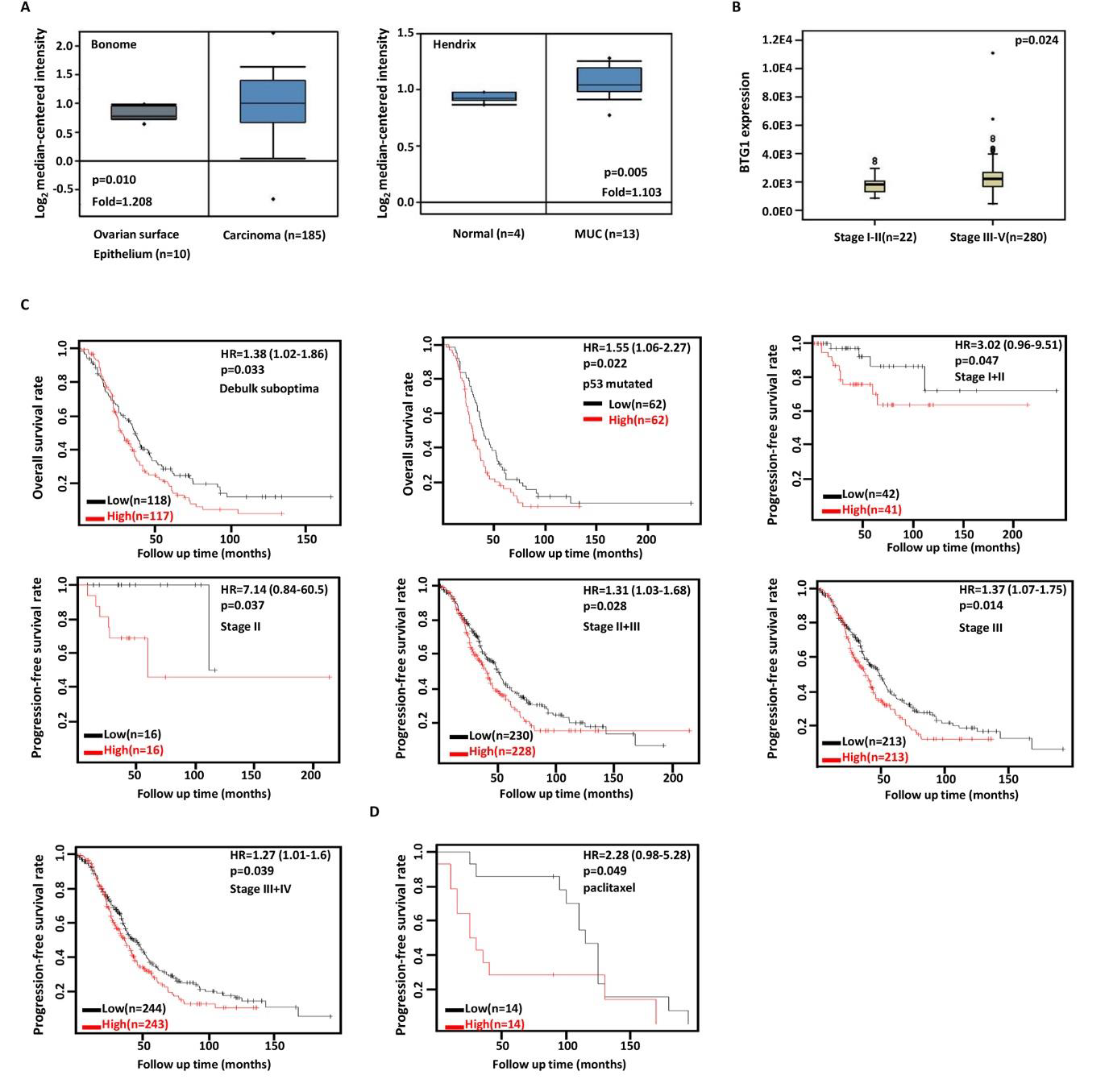
Figure 4: The clinicopathological and prognostic significances of BTG1 mRNA expression in ovarian cancer
Bonome’s and Hendrix’s datasets were employed for bioinformatics analysis to observe BTG1 expression in ovarian cancer. A lower BTG1
expression was detectable in ovary than that in ovarian carcinoma or mucinous adenocarcinoma (MUC) respectively (A, p<0.05). TCGA database
showed that BTG1 was more expressed in stage III-IV than I-II cancer (B, p<0.05). The correlation between expression and overall, or postprogression
survival rate of the patients with ovarian cancer, even stratified by different clinicopathological parameters (C, p<0.05). HR, hazard ratio.
Conclusion
BTG1 overexpression suppressed proliferation, migration and invasion, and induced apoptosis and cell cycle arrest of hepatocellular, thyroid, nasopharyngeal, esophageal, breast, and non-small cell lung cancer cells with down-regulated expression of Cyclin D1, Bcl-2, and MMP-9 [21-26]. In ovarian cancer, BTG1 expression caused lower growth rate, high cisplatin sensitivity, G1 arrest, apoptosis, and decreased migration and invasion by down-regulating the expression of PI3K, PKB, Bcl-xL, survivin, VEGF, and MMP-2 [27]. The chemosensitivity of BTG1 transfectants to paclitaxel, cisplatin, MG132 or SAHA was positively correlated with its apoptotic induction of colorectal cancer cells [18]. BTG1 overexpression suppressed tumor growth and lung metastasis of gastric and colorectal cancer cells by inhibiting proliferation and enhancing autophagy and apoptosis in xenograft models [17,18]. Taken together, BTG1 should be used as a molecular target for cancer gene therapy. Jung et al. [28] found that BTG1 expression was lower in colorectal cancer than control, and in metastatic than primary cancer, due to the hypermethylation of BTG1 promoter. BTG1 expression was decreased in hepatocellular, thyroid, nasopharyngeal, esophageal, breast, colorectal, and non-small cell lung cancers, and negatively associated with aggressive behaviors [20-26]. Decreased BTG1 expression in gastric cancer was positively correlated with depth of invasion, lymphatic and venous invasion, lymph node metastasis, TNM staging and worse prognosis [16], but the lower BTG1 expression in ovarian cancer was positively correlated with FIGO staging [25]. Here, it was found that BTG1 expression was lower in gastric cancer than normal mucosa in line with both Kanda’s and our reports [17,29], but versa for pulmonary squamous cell carcinoma, breast invasive ductal cancer, and ovarian cancer, different from the other results [24-26]. Reportedly, the downregulated BTG1 expression is positively correlated with its promoter methylation in colorectal, gastric, and ovarian cancers [17,29 &30]. Moreover, BTG1 expression was positively correlated with dedifferentiation and histological grading and of gastric cancer, and TNM staging of ovarian cancer, but negatively associated with lymph node metastasis and TNM staging of breast cancer. BTG1 expression was higher in pulmonary squmaous cell carcinoma than adenocarcinoma, which provided another evidence that squamous cell carcinoma but not adenocarcinoma showed higher BTG1 expression than normal tissue. These findings suggested that aberrant BTG1 expression was positively correlated with carcinogenesis, histogenesis and subsequent progression. The paradoxical data in our and other data may be due to sample selection, different methodologies, and tissue specificity. Kanda et al. [29] reported that downregulation of BTG1 mRNA in gastric cancer was positively associated with shorter disease-specific and recurrence-free survival of the patients with gastric cancer as an independent prognostic factor. BTG1 expression was adversely linked to poor prognosis of the patients with hepatocellular, thyroid, nasopharyngeal, esophageal, breast, or non-small cell lung cancer [22-26]. According to Kaplan-Meier plotter, we found that BTG1 expression was negatively correlated with favorable prognosis of the gastric, lung or ovarian cancer patients, but versa for breast cancer patients. The correlation between BTG1 expression and prognosis of the cancer patients did not parallel with the alteration in BTG1 expression in cancer tissues. Additionally, TCGA data showed the same results about the prognostic significance of BTG1 expression to KM plotters in gastric and breast cancers although KM plotter is based on cDNA array and TCGA experiment on RNA sequencing. The tissue specificity might account for the phenomenon about the relationship between BTG1 expression and prognosis.
In conclusion, aberrant BTG1 mRNA expression was closely linked to carcinogenesis, cancer aggressiveness, and worse prognosis of the cancer patients in a tissue-specific manner. The limitation of this study is not to verify the results from Oncomine, TCGA and KM plotter datasets using real-time RT-PCR, even after laser capture dissection.
Acknowledgement
This study was supported by National Natural Scientific Foundation of China (81472544; 81672700).
Conflict of Interest
The authors have declared that no competing interests exist.
References
- Busson M, Carazo A, Seyer P, Grandemange S, Casas F, et al. (2005) Coactivation of nuclear receptors and myogenic factors induces the major BTG1 influence on muscle differentiation. Oncogene 24(10): 1698-1710.
- Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR (1996) The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem 271(25): 15034-15044.
- Bogdan JA, Adams Burton C, Pedicord DL, Sukovich DA, Benfield PA, et al. (1998) Human carbon catabolite repressor protein (CCR4)-associative factor 1: cloning, expression and characterization of its interaction with the B-cell translocation protein BTG1. Biochem J 336(Pt2): 471-481.
- Prévôt D, Voeltzel T, Birot AM, Morel AP, Rostan MC, et al. (2000) The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem 275(1): 147-153.
- Corjay MH, Kearney MA, Munzer DA, Diamond SM, Stoltenborg JK (1998) Antiproliferative gene BTG1 is highly expressed in apoptotic cells in macrophage-rich areas of advanced lesions in Watanabe heritable hyperlipidemic rabbit and human. Lab Invest 78(7): 847-858.
- Nahta R, Yuan LX, Fiterman DJ, Zhang L, Symmans WF, et al. (2006) B cell translocation gene 1 contributes to antisense Bcl-2-mediated apoptosis in breast cancer cells. Mol Cancer Ther 5(6): 1593-1601.
- Liu C, Tao T, Xu B, Lu K, Zhang L, et al. (2015) BTG1 potentiates apoptosis and suppresses proliferation in renal cell carcinoma by interacting with PRMT1. Oncol Lett 10(2): 619-624.
- Zhang H, Tang J, Li C, Kong J, Wang J, et al. (2015) MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett 356(2 Pt B): 781-790.
- Nan YH, Wang J, Wang Y, Sun PH, Han YP, et al. (2016) MiR-4295 promotes cell growth in bladder cancer by targeting BTG1. Am J Transl Res 8(11): 4892-4901.
- Zhang SQ, Yang Z, Cai XL, Zhao M, Sun MM, et al. (2017) miR-511 promotes the proliferation of human hepatoma cells by targeting the 3'UTR of B cell translocation gene 1 (BTG1) mRNA. Acta Pharmacol Sin 38(8): 1161-1170.
- He C, Yu T, Shi Y, Ma C, Yang W, et al. (2017) MicroRNA 301A promotes intestinal inflammation and colitis-associated cancer development by inhibiting BTG1. Gastroenterology 152(6): 1434-1448.
- Zhao X, Chen GQ, Cao GM (2019) Abnormal expression and mechanism of miR-330-3p/ BTG1 axis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 23(16): 6888-6898.
- Li E, Han K, Zhou X (2019) microRNA-27a-3p down-regulation Inhibits Malignant Biological Behaviors of Ovarian Cancer by Targeting BTG1. Open Med (Wars) 14: 577-585.
- Kim M, Jo H, Kwon Y, Kim Y, Jung HS, et al. (2020) Homoharringtonine inhibits allergic inflammations by regulating NF-κB-miR-183-5p-BTG1 Axis. Front Pharmacol 11: 1032.
- Su C, Huang DP, Liu JW, Liu WY, Cao YO (2019) miR-27a-3p regulates proliferation and apoptosis of colon cancer cells by potentially targeting BTG1. Oncol Lett 18(3): 2825-2834.
- Zhu R, Li W, Xu Y, Wan J, Zhang Z (2015) Upregulation of BTG1 enhances the radiation sensitivity of human breast cancer in vitro and in vivo. Oncol Rep 34(6): 3017-3024.
- Zheng HC, Li J, Shen DF, Yang XF, Zhao S, et al. (2015) BTG1 expression correlates with pathogenesis, aggressive behaviors and prognosis of gastric cancer: a potential target for gene therapy. Oncotarget 6(23): 19685-19705.
- Zhao S, Chen SR, Yang XF, Shen DF, Takano Y, et al. (2020) BTG1 might be employed as a biomarker for carcinogenesis and a target for gene therapy in colorectal cancers. Oncotarget 8(5): 7502-7517.
- Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, et al. (2016) Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7(31): 49322-49333.
- Zhu Y, Qiu P, Ji Y (2014) TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods 11(6): 599-600.
- Sun GG, Lu YF, Cheng YJ, Yang CR, Liu Q, et al. (2014) Expression of BTG1 in hepatocellular carcinoma and its correlation with cell cycles, cell apoptosis, and cell metastasis. Tumor Biol 35(12): 11771-11779.
- Lu YF, Sun GG, Liu Q, Yang CR, Cheng YJ (2014) BTG1 expression in thyroid carcinoma: diagnostic indicator and prognostic marker. Int J Oncol 45(4): 1574-1582.
- Sun GG, Wang YD, Cheng YJ, Hu WN (2014) The expression of BTG1 is downregulated in nasopharyngeal carcinoma and possibly associated with tumour metastasis. Mol Biol Rep 41(9): 5979-5988.
- Sun GG, Wang YD, Cheng YJ, Hu WN (2014) BTG1 underexpression is an independent prognostic marker in esophageal squamous cell carcinoma. Tumor Biol 35(10): 9707-9716.
- Sheng SH, Zhao CM, Sun GG (2014) BTG1 expression correlates with the pathogenesis and progression of breast carcinomas. Tumor Biol 35(4): 3317-3326.
- Sun GG, Lu YF, Cheng YJ, Hu WN (2014) The expression of BTG1 is downregulated in NSCLC and possibly associated with tumor metastasis. Tumor Biol 35(4): 2949-2957.
- Zhao Y, Gou WF, Chen S, Takano Y, Xiu YL, et al. (2013) BTG1 expression correlates with the pathogenesis and progression of ovarian carcinomas. Int J Mol Sci 14(10): 19670-19680.
- Jung YY, Sung JY, Kim JY, Kim HS (2018) Down-regulation of B-Cell Translocation Gene 1 by promoter methylation in colorectal carcinoma. Anticancer Res 38(2): 691-697.
- Kanda M, Oya H, Nomoto S, Takami H, Shimizu D, et al. (2015) Diversity of clinical implication of B-cell translocation gene 1 expression by histopathologic and anatomic subtypes of gastric cancer. Dig Dis Sci 60(5): 1256-1264.
- Kim JY, Do SI, Bae GE, Kim HS (2017) B-cell translocation gene 1 is downregulated by promoter methylation in ovarian carcinoma. J Cancer 8(14): 2669-2675.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.