Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Altering Trichomonas Vaginalis Codons Enhances Expression of The Trichomonad P270 Repeat Element in Escherichia Coli
*Corresponding author: JF Alderete, School of Molecular Biosciences, MC7520, College of Veterinary Medicine, Washington State University, Pullman, WA 99164.
Received: October 05, 2021; Published: November 02, 2021
DOI: 10.34297/AJBSR.2021.14.002024
Abstract
The immunogenic protein of the Trichomonas vaginalis isolate NYH 286 called P270 (270-kDa) has 19 tandemly repeated elements (TRE), and each RE is 333-bp, which encodes for a protein of 11,770.90 daltons. Isolate NYH 286 trichomonads are infected with a dsRNA virus (TVV+), and TVV+ organisms undergo phenotypic variation between surface versus cytoplasmic placement of P270. These TVV+ trichomonads are heterogeneous with fluorescent and non-fluorescent parasites by indirect immunofluorescence using the monoclonal antibody (MAb) C20A3 as a probe. The p270 gene is single copy based on restriction enzyme analysis, and partial restriction using HindIII reveals a ladder pattern expected of TREs. Each TRE has the DREGRD epitope detected by MAb C20A3. As the cDNA encoding a protein of ~14-kDa within which is the ~11.8-kDa RE is poorly expressed by recombinant E. coli (rE.coli) the hypothesis was tested that the T. vaginalis codons for arginine (R) and glycine (G) translated by minor E. coli tRNA species decreased the synthesis of the RE. Therefore, the five codons of R109, R112, R116, G119, and G128 were altered with synonymous codons used by E. coli. The synthesis of the RE of the original plasmid (pORIG) expressed in rE. coli was then compared with rE. coli harboring the plasmid with less numbers of codons (called pLESS) used by the minor E. coli tRNAs. The control rE. coli with plasmid p21.b without insert, pORIG and pLESS all had similar growth kinetics, and bacterial lysates harvested at different times were evaluated by immunoblot for synthesis of the RE. The data show that the rE. coli with pORIG synthesized the RE only between 2-hours (h) and 8-h of growth. In contrast, rE. coli with pLESS permitted synthesis of the RE during the 30-h of bacterial growth. These data show for the first time the role of codon usage for expression of T. vaginalis proteins in rE. coli.
Keywords: Codon Usage; Escherichia Coli Codons; Gene Expression; P270; Silent Codon Changes; Tandemlyrepeated Element; Trichomonas Vaginalis; Transfer RNA.
Abbreviations: amp: ampicillin; bp: base pairs; coli: Escherichia coli; kb: kilobases; Mab: monoclonal antibody; Mr: electrophoretic mobility; MW: molecular weight; NC: nitrocellulose; P270: phenotypically varying high Mr protein; SDS-PAGE: sodium dodecylsulfate polyacrylamide gel electrophoresis; TRE: tandemly repeated element; tRNA: transfer RNA
Introduction
Trichomonas vaginalis remains the number one, non-viral sexually transmitted infection (STI) worldwide [1,2]. Indirect immunofluorescence (IF) using monoclonal antibody (MAb) C20A3 revealed naturally occurring isolates of T. vaginalis with trichomonads that were heterogeneous for the surface expression of a 270-kDa protein called P270 [3-5]. Flow cytometry experiments revealed that fluorescent organisms became non-fluorescent and vice-versa during in vitro batch culture [3], and this property of phenotypic variation was found to occur only for isolates of T. vaginalis harboring a dsRNA virus (TVV+) [6,7]. This finding led to the discovery of two naturally occurring T. vaginalis isolates types defined as Type I without dsRNA virus (TVV-) and Type II (TVV+) [6]. Of particular interest is the finding that extended batch cultivation of TVV+ parasites abort the virus thus creating TVV- like isogenic isolates which have lost the ability to undergo phenotypic variation [6-8], suggesting an important role for the virus in regulating surface placement of P270.
The large Mr P270 protein from representative TVV+ phenotypically varying trichomonads was found to have an internal region comprised of tandemly repeated element (TRE) sequences [9,10], and each TRE had the immunodominant DREGRD amino acid sequence detected by the MAb C20A3 [10-12]. Although the function of P270 remains undefined, recent work combining IF and immunoelectron microscopy has shown the existence of P270 on both the surface and inside the cytoplasm and peripheral vacuoles within the TVV+ parasites [8]. Importantly, P270 is stably conserved among fresh clinical isolates, which suggests an important function for P270 in the biology of T. vaginalis.
Early attempts to synthesize the RE as a recombinant protein by E. coli was met with limited success [9]. Expression of the RE was suboptimal, which did not permit for structure-function studies. In this paper, the hypothesis that the poor synthesis of the RE in rE. coli is due to the presence of T. vaginalis codons that are used by minor tRNAs of E. coli [13] was tested. Four arginine and two glycine codons were changed in the original cDNA encoding the RE (pORIG) with synonymous codons used by rE. coli. This resulted in a modified plasmid (called pLESS) with less numbers of codons that use minor E. coli tRNAs. Data presented here show tje synthesis of the recombinant RE throughout the extended growth of rE. coli with pLESS compared with rE coli harboring the plasmid pORIG. These findings suggest that codon usage within trichomonad genes is an important variable that influences the kinetics and amount of recombinant protein produced rE. coli. The significance of our observations is discussed.
Methods
Screening of the transformants
Both blue-white and immunological screening of bacterial colonies using the MAb C20A3 [3,4] were performed by standard procedures [9]. White, immunoreactive clones were further evaluated through double restriction of purified plasmids (Qiagen Inc., Valencia, CA USA). Recombinant plasmids were verified by sequencing. Secondary structures of recombinant mRNAs were analyzed by PC/GENE (IntelliGenetics, Atlanta, GA USA).
pORIG and pLESS plasmids
Phagemid vector pcDNAII (Thermo Fisher Scientific, Waltham, MA USA) was employed for cDNA cloning. Subcloning of trichomonad cDNA required PCR amplification using the original cDNA clone [9] as template. Unidirectional ligation of the original cDNA into the plasmid (called pORIG) was assured by using a sense primer (ORIG) (5’-TTGAATTCCGGGATAACGTTAGA-3’) designed to introduce an EcoRI restriction at the 5’ site and an antisense primer (labeled orig) (5’-TTGATATCCCCTTGTTGTGCTGCGCG-3’) to introduce an EcoRV restriction at the 3’ site.
To decrease the number of codons in the original cDNA that use minor tRNAs for the pLESS plasmid for expression, the same orig sense primer was used, and a new antisense primer (labeled less) (5’-TTGATATCGCCTTGTTGTGCTGCGCGTAATGTGTCGCCTTTACTGCGAACGTTATCGCGACCTTCTCGAT-3’) was made to replace five codons translated by minor E. coli tRNA species [13]. Three arginine codons (R109, R112, and R116)and two glycine codons (G119, and G128) of the original cDNA (Figure 1) [9] were changed with synonymous codons to replace the T. vaginalis codons. The synonymous codons were selected to avoid introducing secondary mRNA structural and translation initiation issues, which were analyzed as described above by PC/GENE. Hot start PCR amplification (Clontech Laboratories, Mountain View, CA USA) with three-step cycles was performed with the Ta individually set for each pair of primers. The amplification products were purified (Wizard PCR Preps, Promega, Madison, WI USA), digested with EcoRI and EcoRV restriction enzymes, and ligated into the EcoRI/EcoRV-directed vector site. Transformation into the INVαF’ host strain was performed as recommended by the manufacturer.
Recombinant E. coli and expression of the P270 repeat element (RE)
Recombinant E. coli (rE. coli) INVαF' (Thermo Fisher Scientific) with the plasmids pORIG and pLESS (Figure 1) as well as control E. coli with p21.b were grown in Luria-Bertani (LB) medium and/or on LB agar plates supplemented with 60µg/ml of ampicillin (amp) as described recently [14-16]. Twenty milliliters of LB medium were then inoculated with equal amounts of bacteria and incubated at 37°C for up to 32-h on a shaker at 250rpm. At 2-h intervals, an aliquot of each culture with equal OD600 was centrifuged, and pellets were frozen at -20°C until used for electrophoresis and immunoblotting as described below. Also, the plasmids from an equal volume of each rE. coli with identical OD600 were purified [9,14,15]. Both purified plasmids and cDNAs were quantitated after electrophoresis in agarose gels to ensure the rE. coli possessed equivalent amounts of pORIG and pLESS plasmid DNA. This was done to be able to accurately compare the P270 REs synthesized by rE. coli.
Sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on lysates derived from frozen pellets of each rE. coli with the p21.b control plasmid and the pORIG and pLESS plasmids obtained above. Pellets were thawed with electrophoresis dissolving buffer, as before [9,12,14]. Stained gels for rE. coli was obtained for time points of 2-h up to 30-h to insure equal amounts of protein were added to lanes of all time points for SDS-PAGE and subsequent immunoblotting onto nitrocellulose. Duplicate gels after SDS-PAGE were transferred onto nitrocellulose for probing with the MAb C20A3 that detects the TRE DREGRD (Figure 2) conserved within the RE as described before and using established protocols [9,10,12,14-16].

Figure 1: Nucleotide and amino acid sequences of the cDNAs of plasmid pORIG as in Figure 1 and of plasmid pLESS with altered codons. The designation pLESS denotes a decrease in codons that utilize minor tRNAs in E. coli. The altered codons for R109, R112, R116, G119, and G128 are in red, underlined and boxed in red to reflect the translation by alternative tRNA species by E. coli. The replaced synonymous codons retain the same amino acids. The DREGRD epitope as above for Figure 2 is also highlighted and boxed in red.

Figure 2: Open reading frame and amino acid sequence of the cDNA called pORIG of the P270 TRE, as before [9]. The two brackets shown in red highlight the 49 nucleotides (17 amino acids) that are repeated within the cDNA. The amino acid DREGRD boxed in red is the tandemly repeated epitope within the p270 gene that is detected by the monoclonal antibody (MAb) C20A3 [8,9,11]. The restriction sites of EcoRI and EcoRV for cloning purposes are shown at the 5' and 3' ends of the cDNA, respectively.
Results
The p270 gene and tandemly repeated elements (TREs)
As in earlier reports for a T. vaginalis Type II (TVV+) isolate [10,11] the organization of the p270 gene and TREs for the long-term grown NYH 286 isolate were characterized. Figure 3 (part a) illustrates the p270 gene with the internal large TRE of 333-bp defined by the restriction enzyme HindIII [9,10] that encodes for a protein of ~11.8-kDa, consistent with the p270 gene of a fresh clinical isolate [10,11]. Parts b and c are two different representative experiments of T. vaginalis DNA digested with EcoRI, which shows the p270 gene is single copy consistent with an earlier report [10]. Digestion to completion with HindIII shows a single band of the expected size for part b (lane 1) and part c (lanes 1a through 1c). Partial digestion with HindIII shows the ladder pattern as expected for a repeat element within the p270 gene [12].
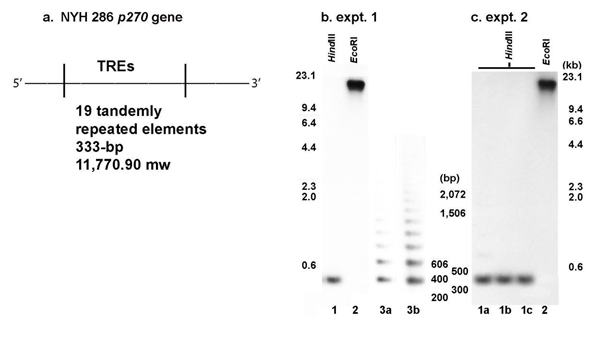
Figure 3: The T. vaginalis NYH 286 isolate p270 gene (part a) and restriction enzyme digestion of the purified trichomonad DNA (part b). Part a presents the organization of the p270 gene within which are the 19 tandemly repeated elements (TREs) of 333-bp defined by HindIII and that encode for a RE of ~11.8-kDa in size. This gene organization is consistent with an earlier published report [11]. Part b presents the complete restriction enzyme digestion of DNA with EcoRI (expt. 1 and 2, lanes 2). The complete digestion with HindIII is shown in lane 1 for expt. 1 and lanes 1a through 1c for expt. 2. Complete digestion for the two representative experiments was performed identically. For both experiments, the expected large size of the p270 gene of ~24-kb in size for EcoRI-digested DNA and the 333-bp size for HindIII-digested DNA were obtained, as evidenced by the size markers and consistent with earlier reports [9-11]. Lanes 3a and 3b of expt. 1 is of partial HindIII restriction to illustrate the 333-bp TRE ladder compared to the full digestion single band end product as shown in lane 1 of expt. 1 and lanes 1a-1c of expt. 2.
Plasmids pORIG and pLESS encoding the TRE and codon usage
Figure 2 presents the 384 nucleotide RE of the p270 gene that is withing the 396 nucleotide cDNA described before [9]. Flanking the DNA is an EcoRI and EcoRV restriction sites at the 5' and 3' ends, respectively, used for cloning into pORIG. The brackets in red shows the repeated 49 nucleotide (17 amino acids) that flank the cDNA, and the immunodominant epitope DREGRD detected by MAb C20A3 is boxed in red. This original 384bp cDNA (pORIG) encodes for a protein of ~14.2-kDa as described before [9].
Comparison of pORIG with the pLESS cDNA having five less T. vaginalis codons that use minor tRNAs of E. coli is shown in Figure 1. The modified codons R109, R112, R116, G119, and G128 are in red, underlined and boxed in red. As for Figure 2, the DREGRD epitope is boxed in red. In a separate experiment, the codons R5, G8, and G17 in pLESS were also modified, but these changes did not increase the amount of RE that is synthesized and are not included here.
Synthesis of the P270 TRE
The synthesis of the p270 RE first required showing that the growth kinetics of each rE. coli with the respective plasmids (p21.b, pORIG and pLESS) were similar, and the growth curves of rE. coli is shown in Figure 4. Each rE. coli reached similar OD600 densities. This was preparatory to performing SDS-PAGE and immunoblotting using as probe the MAb C20A3. The rE. coli was harvested at 2-h or 4-h intervals until 30-h of growth.
Figure 5 presents the Coomassie brilliant blue-stained gels (labeled a) and immunoblots (labeled b) after SDS-PAGE of total proteins of E. coli harboring the plasmids p21.b control (part A), pORIG (part B) and pLESS (part C). For each rE. coli throughout the growth period, stained gels of total E. coli proteins were identical, showing that lanes for each time period were loaded with the same amount of rE. coli protein. Not unexpectedly, the immunoblot for control rE. coli with p21.b was unreactive with the MAb C20A3 as evidenced by the absence of any protein band (part A). Ponceau-staining of the nitrocellulose showed equal amounts of total rE. coli proteins as the stained gels. For the rE. coli with pORIG ( part B), the MAb C20A3 reacted with the RE protein of the expected ~14-kDa in size but only for the 2-h to 8-h time period. On the other hand, the nitrocellulose with proteins of rE. coli with pLESS (part C) showed the RE protein throughout the growth curve up to 30-h. These data suggests strongly that the changes in the codons to those utilized by E. coli resulted in more effective synthesis of the RE. For rE. coli with pORIG or pLESS, there was no evidence of overexpression of P270 in stained gels.
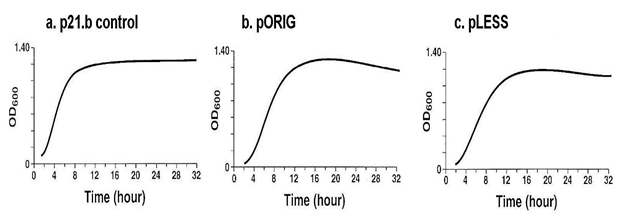
Figure 4: The growth curves of respective E. coli control with plasmid without insert (a, p21.b), original plasmid (b, pORIG) and plasmid with modified codons (c, pLESS). Each recombinant E. coli reached similar OD600 densities, and recordings were carried out every 2-h until 12-h after which bacterial growth densities were recorded every 4-h up until 32-h. This was necessary because immunoblotting after SDS-PAGE of total E. coli proteins was carried out up to 30-h for expression of the P270 cDNA encoding the RE.
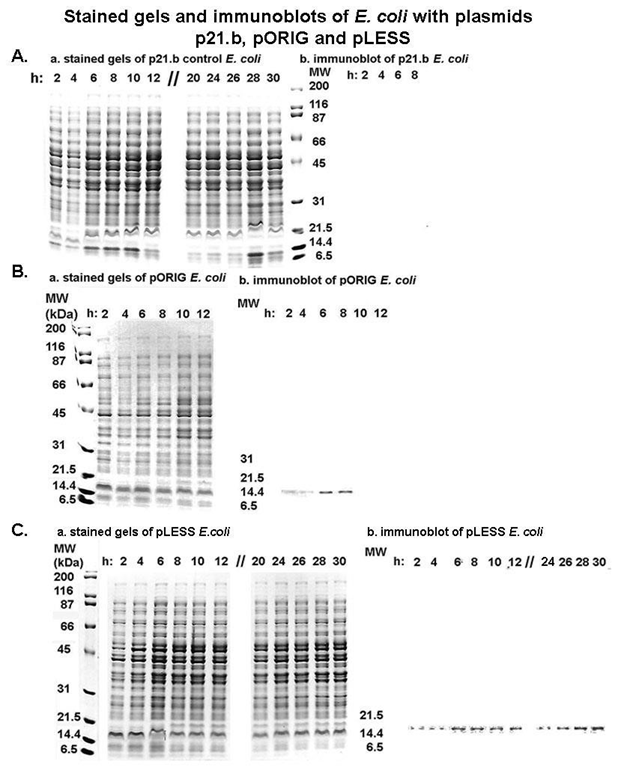
Figure 5: Coomassie brilliant, blue-stained gels (labeled a) and immunoblots (labeled b) after SDS-PAGE of total proteins of E. coli harboring the plasmids p21.b control (part A), pORIG (part B) and pLESS (part C). For experiments with the three plasmids, the stained gels show that equal amounts of protein were added to wells for each time point for each rE. coli (Part A, Part B and Part C, respectively). Part A. Recombinant E. coli with control p21.b was grown for up to 32-h. E. coli were harvested at the indicated times and total proteins were electrophoresed and stained. Duplicate gels were immunoblotted onto nitrocellulose membranes and probed with the P270 TRE MAb C20A3 that detects the epitope DREGRD (Figures 2 and 3). The blot for only 2-h through 8-h is shown, although no proteins were detected at all time points. Part B. Same as part A except using rE. coli with pORIG. Immunoblots with MAb C20A3 showed a protein band with the expected MW of ~14-kDa for the cDNA encoding the TRE, as before [9,13]. Only up to 12-h for the stained gel is presented as no protein was evident after 8-h in the immunoblot. Part C. Same as part A except with rE. coli with pLESS. The immunoblot probed with MAb C20A3 shows the presence of recombinant RE throughout the 30-h growth of rE. coli beginning at 2-h through 30-h. For E. coli with pORIG or pLESS, there was no evidence of overexpression of P270 in stained gels. The experiments were performed no less than on three different occasions. Importantly, for each of the nitrocellulose blots, Ponceau staining showed total proteins as seen for the stained gels presented in part a.
Discussion
In this report we wanted to test the hypothesis that the poor synthesis of the p270 tandemly repeated element was the result of T. vaginalis using codons for arginine and glycine translated by rare E. coli tRNAs [13,17-21]. This is the first time that rE. coli expressing a RE of p270 from pLESS with three altered arginine and two altered glycine codons is shown to be synthesized throughout the bacterial growth kinetics compared with pORIG-expressing rE. coli (Figure 5). Indeed, our observations reinforced the idea that the presence of rare codons in the cloned trichomonad cDNA directly affects amounts of recombinant protein synthesized in rE. coli. The examination of 29,845 codons from 80 partial and complete sequences of proteincoding T. vaginalis genes in GenBank further revealed that 2% of leucine, 4% of isoleucine, 11% of glycine, and 25% of arginine residues were encoded by codons rarely used in E. coli [13,17-21]. Thus, it is conceivable that RE gene expression in pORIG is influenced by different rates of translation resulting from the amounts of minor tRNAs.
The level of gene expression depends on amount and translational frequency of full-length mRNA [21]. Both pORIG and pLESS cDNAs were ligated into identical sites of lacZ gene, and consequently expression of both proteins was under the control by the same promoter. Importantly, the vector does not express the lac repressor, further affirming that both cDNAs were under the control of equal elements within the plasmid and cDNA. Two classes of protective elements have been shown to stabilize mRNAs in E. coli. One class of elements are sequences in the 5'-UTRs, and the second group includes stem-loop structures from the 3'-UTRs [19]. It is equally noteworthy that as no sequence changes were done within the 5'-UTRs of the cDNAs, evaluation of the 3'-UTR regions of the mRNAs from pORIG and pLESS did not detect any differences in the secondary structure. Furthermore, although silent codon changes influence mRNA stability via variations of ribosome traffic [21,22], it has been established that abundance of different tRNA species varies with the growth rates of E. coli [23]. Therefore, the data on RE synthesis in pLESS versus pORIG may be explained by numerous variables as mentioned here. This notwithstanding, our work suggests that the use by T. vaginalis of tRNAs that are minor in E. coli is likely to negatively affect successful stable expression of recombinant trichomonad proteins. Our results may have bearing on the efficient synthesis of this RE and other trichomonad proteins in rE. coli, and synthesis of the T. vaginalis proteins is a prerequisite first step in structure-function characterization of virulence factors and, therefore, of proteins important to the hostparasite interrelationship.
Conclusion
The ability to express proteins of T. vaginalis in rE. coli is preparatory for structure-function studies that permit understanding of the role of such proteins in virulence and mechanisms of disease pathogenesis for this STI caused by this ancient protist. This work examined for the first time the role of codons used by trichomonads in the poor expression of proteins in rE. coli. Future successful synthesis of heretofore poorlyexpressed T. vaginalis virulence factors in rE. coli via alteration of codons will contribute to our overall understanding of the biology of this STI agent and of the ability of this organism to infect human hosts.
Acknowledgement
This study was supported by Public Health Service grants AI- 39803 and AI-43940 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health while JFA was at The University of Texas Health Science Center at San Antonio. As per instructions by the International Committee of Medical Journal Editors on “Who is an Author?,” (http://www.icmje.org/recommendations/browse/roles-andresponsibillities/definingthe-role-of-authors-and-contributors.html), I want to acknowledge Oxana Musatatova and Jean Engbring who generated the data under my supervision in my laboratory while at The University of Texas Health Science Center at San Antonio.
References
- World Health Organization (1995) An overview of selected curable sexually transmitted diseases. WHO Global Programme on AIDS Report.
- Hobbs MM, Sena Ac, Swygard H, Schwebke Jr (2008) Trichomonas vaginalis and trichomoniasis. Holmes KK, Sparling PF, Stamm WE, Piot P, Wasserheit JN, et al. Eds. Sexually Transmitted Diseases, McGraw-Hill Medical, New York, USA.
- Alderete JF, Kasmala L, Metcalfe E, Garza GE (1986) Phenotypic variation and diversity among Trichomonas vaginalis isolates and correlation of phenotype with trichomonal virulence determinants. Infect Immun 53(2): 285-293.
- Alderete JF, Demes P, Gombosova A, Valent M, Janoska A, et al. (1987) Phenotypes and protein-epitope phenotypic variation among fresh isolates of Trichomonas vaginalis. Infect Immun 55(5): 1037-1041.
- Alderete J F, Suprun Brown L, Kasmala L (1986) Monoclonal antibody to a major surface immunogen differentiates isolates and subpopulations of Trichomonas vaginalis. Infect Immun 52(1): 70-75.
- Wang A, Wang CC, Alderete JF (1987) Trichomonas vaginalis phenotypic variation occurs only among trichomonads infected with the double-stranded RNA virus. J Exp Med 166(1): 142-150.
- Khoshnan A, Alderete JF (1994) Trichomonas vaginalis with a double-stranded RNA virus has upregulated levels of phenotypically variable immunogen mRNA. J Virol 68(6): 4035-4038.
- Alderete JF (2021) Localization of the phenotypically varying P270 protein on dsRNA virus-positive and negative Trichomonas vaginalis Amer J Biomed Sci Res 14(2): 199-209.
- Dailey DC, Alderete JF (1991) The phenotypically variable surface protein of Trichomonas vaginalis has a single, tandemly repeated immunodominant epitope. Infect Immun 59(6): 2083-2088.
- Musatovova O, Alderete JF (1999) The Trichomonas vaginalis phenotypically varying p270 immunogen is highly conserved except for numbers of repeated elements. Microb Pathogen 27(2): 93-104.
- Musatovova O, Alderete JF (1998) Molecular analysis of the gene encoding the immunodominant phenotypically varying p270 protein of Trichomonas vaginalis. Microb Pathog 24(4): 223-239.
- Alderete JF, Neale KA (1989) Relatedness of structures of a major immunogen in Trichomonas vaginalis Infect Immun 57(6): 1849-1853.
- Ikemura T (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translation system. J Mol Biol 151(3): 389-409.
- Arroyo R, Engbring J, Nguyen J, Musatovova O, Alderete JF, et al. (1995). Characterization of cDNAs encoding adhesin proteins involved in Trichomonas vaginalis Arch Med Res 26(4): 361-369.
- Alderete JF, O Brien JL, Arroyo R, Engbring JA, Musatovova O, et al. (1995) Cloning and molecular characterization of two genes encoding adhesion proteins involved in Trichomonas vaginalis Mol Microbiol 17(1): 69-83.
- Alderete JF (2021) Recombinant protein of immunogenic metabolic enzyme epitopes of Trichomonas vaginalis are common to humans and microorganisms. Amer J Biomed Sci Res 13(6): 630-638.
- Grosjean H, Fiers W (1982) Preferential codon usage in prokaryotic genes: the optimal condon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene 18(3): 199-209.
- Zahn K (1996) Overexpression of an mRNA dependent on rare codons inhibits protein synthesis and cell growth. J Bacteriol 178(10): 2926-2933.
- Makrides SC (1996) Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev 60(3): 512-538.
- Chen GT, Inouye M (1994) Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Gen Develop 8(21): 2641-2652.
- Deana A, Ehrlich R, Reiss C (1996) Synonymous codon selection controls in vivo turnover and amount of mRNA in Escherichia coli bla and ompA genes. J Bacteriol 178(9): 2718-2720.
- Deana A, Ehrlich R, Reiss C (1998) Silent mutations in the Escherichia coli ompA leader peptide region strongly affect transcription and translation in vivo. Nucleic Acids Res 26(20): 4778-4782.
- Emilsson V, Näslund AK, Kurland CG (1993) Growth-rate-dependent accumulation of twelve tRNA species in Escherichia coli. J Mol Biol 230(2): 483-491.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.