Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
New Quantitative PCR Technology for Screening Human Papillomavirus Test – Identification of 25 Genotypes: A Molecular Epidemiology and Cross- Sectional Study
*Corresponding author: Thompson Bonilla María del Rocío, Genomic Medicine Laboratory, “1o Octubre” Regional Hospital, The Security Institute and Workers’ Social Services of the State, Mexico City, Mexico.
Received: January 26, 2022; Published: February 07, 2022
DOI: 10.34297/AJBSR.2022.15.002116
Abstract
Background
A kit for screening HPV-genotypes based on the prevalence of Mexican population there isn´t designed
Objective
To design a new molecular test for human papillomavirus (HPV) screening.
Methods
Molecular epidemiology and cross-sectional study. Multicenter from referral centers. 326 women were included. The primary outcome was designed of a kit for screening HPV-genotypes based on the prevalence found in population. Oligonucleotides for different genotypes were designed. The conditions for quantitative PCR and the pools for low- and high-risk (LR and HR) of HPVgenotypes were established.
Results
With new molecular test we detected 30 HPV-genotypes. A kit with the 25 most prevalent genotypes distributed was designed: four HR genotypes (16, 18, 31, and 59), three LR genotypes (6, 53, and 61), and two pools collectively detected. One pool of another 10 HR genotypes: 33, 35, 39, 45, 51, 52, 56, 58, 66, and 68, and finally a pool of 8 LR genotypes: 11, 40, 43, 44, 55, 70, 73, and 81. The sensitivity of the new molecular test was 97.9% and its specificity was 100% based on the sequencing results.
Conclusion
Our findings provide evidence that the new molecular test for screening of HPV can serve as a better triage test.
Keywords: Human Papillomavirus; Pap Test; Quantitative PCR Technology
Introduction
It is well established that cervical screening based on the identification of low- and high-risk (LR, HR) human papillomavirus (HPV) genotypes can identify more than 95% of precancerous cervical lesions [1-3]. There is evidence that testing for HPV E6/ E7 viral messenger RNA (mRNA) is more specific than molecular tests using amplification of target DNA [1,4-5]. However, present technics cannot identify all HPV-genotypes or have a relatively low specificity for cervical intraepithelial neoplasia [CIN] grade 2 or 3 [CIN2+]) [1,6-7]. The use of primary HR-HPV screening in the cervical cancer programmes has an acceptable sensitivity, specificity, and positive predictive value (PPV), but its sensitivity limits its utility for triage.1 HPV-genotypes 16 and 18 are the genotypes with the highest potential for oncogenesis, and genotypes 31, 33, 45, 52, and 58 explain about 18.5% of malignant neoplasms [1,8-10]. Today, vaccines (tetravalent and nonavalent) have approximately 79- 89% of protection again HPV infection (even cross protection for 31, 33 and 45 genotypes), and 85-95% of potential prevention for vulvar, vaginal, and anal cancers related to this virus [14]. The aim of the current study was to design a new molecular test (care GENETM- HPV test) for HPV screening and compared versus “COBAS” test and Pap test.
Materials and Methods
A multicenter, cross-sectional, and molecular epidemiology study was designed. The subtypes of low- and high-risk HPV genotypes more prevalent were identified in eligible population. A cohort of patients was subjected to two molecular diagnostic tests and Pap test (as the reference test). The data collection was carried out according to a prospective cohort design. All specimens were collected from all levels of health care. We compared the HPV-genotypes identified by care GENETM-HPV test with COBAS system test and Pap test. A diagnostic test evaluation was estimated.
Molecular and Sampling Procedures for Detection of Human Papillomavirus Infection
Two tests were used for a molecular analysis:
i. COBAS system test.
ii. care GENETM-HPV detection kit-I (care GENETM-HPV test).
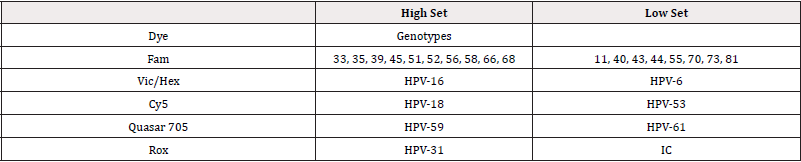
The COBAS system uses an amplification of target DNA by the technics of PCR and nucleic acid hybridization, for detecting a group of 14 HR-HPV genotypes in a single analysis: genotypes 16 and 18 (HPV-16 and HPV-18), and a pool of other HR-HPV genotypes: 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68 [10]. The new test “care GENETM-HPV detection kit-I” employs TaqMan probe qPCR technology for the amplified HPV-DNA detection. The kit was initially designed to detect 33 genotypes, of which 22 are high risk and 11 are low risk. In preliminary tests, 29 genotypes out of 33 possible were detected. Finally, we designed a kit with the 25 most prevalent genotypes, distributed as follows: four HR genotypes (16, 18, 31, and 59), three LR genotypes (6, 53, and 61), and two pools collectively detected. One pool of another 10 HR genotypes: 33, 35, 39, 45, 51, 52, 56, 58, 66, and 68, and finally a pool of 8 LR genotypes: 11, 40, 43, 44, 55, 70, 73, and 81 from crude cervical scraping. Probes for HR-HPV genotypes specific sequences were labelled with the fluorophore Fam (10 HR-HPV genotypes), VIC (HPV-16), Cy5 (HPV-18), ROX (HPV-31), and Quasar705 (HPV-59). Probes for LR-HPV genotypes specific sequence were labelled with the fluorophore FAM (eight LR-HPV genotypes), HEX (HPV-6), Cy5 (HPV- 53), and Quasar 705 (HPV-61). The probe for internal control (IC) was labelled with fluorophore ROX (Table S1). The probe specifically binds to target sequences and fluorescence increases due to the separation of fluorescent dye and quencher by Taq polymerase exonuclease activity during amplification. This test consists of two reaction mixture (with Taq polymerase, dNTP mixture, UDG, and reaction buffer), Mix H (4x Primer/Probe Mixture with HR-HPV specific primer and TaqMan probe), Mix L (4x Prime/Probe Mixture with LR-HPV specific primer and TaqMan probe), PC1 (Positive control for Mix H), PC2 (Positive control for Mix L), and Nuclease-free Water. HPV type-specific signals are detection from fluorescent dye on TaqMan probe, Ct (Cycle threshold) value from qPCR machine represents the relative degree of infection.
Sample Preparation, Storage Condition, and Setting Analysis and Cut Off Value by Qpcr Technology
The sample is human genomic DNA prepared by a genomic DNA isolation kit (QIAamp DSP DNA mini kit or equivalent). Isolated genomic DNA has to be stored below -200C (sealed). It is stable and can be used for 12 months from date of manufacture. The Real-time PCR reaction Mix and PCR amplification steps were done according to manufacturer’s recommendations (Table S2). The analysis setting and acceptance criteria employed also were the recommended by fabricant (Positive: Ct value of signal is 45 or less. Negative: Ct value of signal is not detected according to the fluorescence results, (Table S3)).
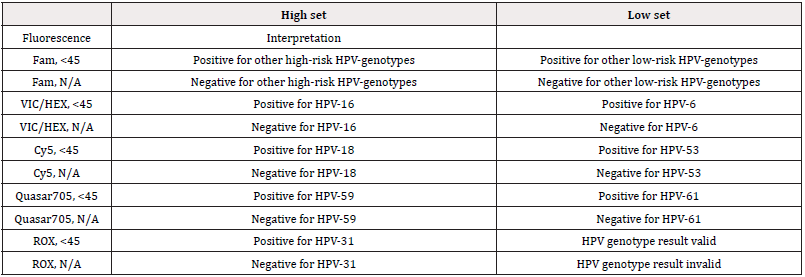
Table S3: Acceptance criteria.
HR: High-risk pool; (Others 10 genotypes): genotype 31, 35, 39, 45, 51, 52, 56, 58, 66, and 68.; LR: Low-risk pool; (Others 8 genotypes): genotypes 11, 40, 43, 44, 55, 70, 73, 81.
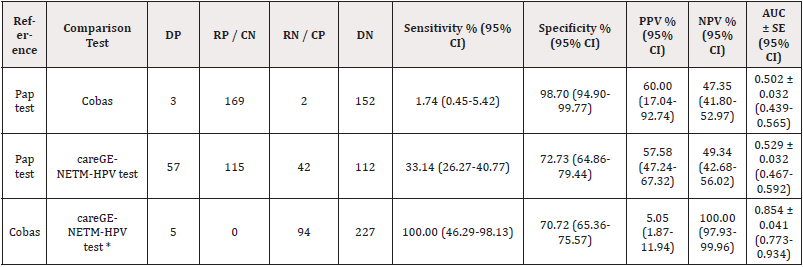
Table 1: Comparison and evaluation of different diagnostic molecular tests for detection of human papillomavirus.
DP: Double positive; RP: Reference-positive; CN: Comparison-negative; RN: Reference-negative; CP: Comparison-positive; DN: Doble negative; PPV: Positive predictive value; NPV: Negative predictive value; %: percentage, CI: Confidence interval; AUC: area under curve. Parameters for calculated the standard error of the area under curve, assumption non-parametric distribution. * Statistical Significance <0.01
Outcome, Study Variables, Statistical Analysis and Ethical Considerations
The primary outcome of the present study was detection of HPV-genotypes. We included age, HPV genotypes and place of residence as study variables. All information was included in a database. Categorical variables were described by both the absolute frequency and percentage with the corresponding 95% confidence interval (95% CI). All categorical variables were compared using chi square test or Fisher exact test as appropriate. The continuous variables were described by mean and standard deviation (SD). To calculate sensitivity, specificity, and predictive positive and negative value, 2 × 2 tables were generated using results from the screening visits and HPV molecular tests. The area under the curve (AUC) was also calculated by a receiver operating characteristic curve (ROC) analysis. Analyses were performed using the total population. All statistical tests were two-sided. A p value < 0.05 (2- sided testing) was considered significant. The Research Ethics Committee, the Research Committee, and the Biosecurity Committee from ISSSTE approved the protocol.
Results
We included a total of 326 women patients with their corresponding samples. The average age of the patients was 42 [1] years old (SD= 11.2, min= 17, max= 84, range= 67, median= 45, interquartile range= 35-51). We include patient´s samples from 6 states (Chiapas, Ciudad de México, Estado de México, Hidalgo, Querétaro, and Tlaxcala). We observed more that 50% samples have low (18.4%) and high (34.4%) grade squamous intraepithelial lesions (LSIL, HSIL). Three samples (from the group of 60 samples with HSIL) had infection with HPV-16 (n= 2) and HPV-18 (n= 1). However, with care GENETM-HPV test we detect a total of 99 positive samples to 30 HPV-genotypes (6, 11, 16, 18, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 55, 56, 58, 59, 61, 62, 66, 67, 68, 69, 70, 73 and 81). The most prevalence genotypes were genotypes 6 (13.2%; 95% CI 9.5-16.9) and 16 (4.6%; 95% CI 2.5-7.1), followed by genotypes 53, 58, 59 (2.8%; 95% CI 1.2-4.6), 31, 61 (2.5%; 95% CI 0.9-4.3), 18, 51, 56 (2.1%; 95% CI 0.6-4.0), and 33 (0.3%; 95% CI 0.0-0.9). Moreover, 35.7% of samples with LSIL (NIC-1) by Pap test are positive to unique (65%) or multiples (35%) infection with HPV-genotypes. Similarly, 28.3% of samples with HSIL (NIC-2) by Pap test are positive to unique (88.2%) or multiples (11.8%) infection with HPV-genotypes (p = 0.1089). Even, 26.2% of negative samples by Pap test are positive to unique (82.4%) or multiples (17.6%) infection with HPV-genotypes. Of 99 positive samples to HPV-genotypes, 74.7% (n= 74) have infection with the presence of 1 genotype of HPV, and 25.3% (n= 25) have co-infection with two or three HPV-genotypes. Three patients had co-infection with three different HPV-genotypes (<18, 51, 56>, <6, 31, 69>, and <18, 31, 39>). The most frequent co-infection observed was the combination between genotype 16, and 61 (12%, three of the 25 patients). From care GENETM-HPV tests, we observed 40 negative samples. Twenty out of the forty-qPCR negative samples showed 150 bp amplicon. Twenty samples with band were sequenced and get the following results:
a. Four Samples were turned out as HR-HPV (Figure 1).
b. Eight samples were turned out as LR-HPV.
c. No sequencing results were obtained from the rest eight samples and were excluded for the accuracy calculation.
Four Samples were turned out as HR-HPV from COBAS system tests. Those four samples are different from the four previous samples of sequencing results. The difference observed might come from the different sensitivity of the two methods. The eight samples excluded for the accuracy calculation in sequencing turned out negative in COBAS system tests. The percentage of infected samples with HPV genotype 33 was 2.2% (5/255), for HPV genotype 31 was 11.5% (12/104), for HPV genotype 58 was 8.6% (22/255) and for HPV genotype 11 was 0.4% (1/255). On the other hand, the percentage of infected samples with HPV-genotype 53 was 2.9% (3/104), and for HPV-genotype 59 was 4.8% (5/104). The ratio of infection of HPV-16 and -18 were 2:1 in simple infections, and 3:1 in co-infections. Generally, it was confirmed that the replaced HPV types are well detected. The sensitivity of the care GENETM-HPV test was 97.9% and its specificity was 100% based on the sequencing results. Data showed that the sensitivity and specificity of the care GENETM-HPV test were 100 and 70.72% compared to COBAS system test, respectively (Table 1). Even more, the area under curve was higher compared to COBAS test (AUC ± SE= 0.854 ± 0.041; 95% CI 0.773-0.934). The kit was designed with the 25 most prevalent genotypes. In relation to low risk-, and high-risk genotypes, we design by each group two pools. We included genotypes 16 and 18 for their oncogenic capacity, with genotypes 31 and 59 (by the distribution observed). Another pool included 10 HR-genotypes (33, 35, 39, 45, 51, 52, 56, 58, 66, and 68). The most prevalence LR-genotypes were included in the same pool (6, 53, and 61). Finally, a pool of 8 LR-genotypes were included: 11, 40, 43, 44, 55, 70, 73, and 81. We included genotype 81 due to the possible epidemiological and public health transcendence.
Discussion
According to Bruni, et al. [9]. the estimated global HPV infection prevailed in women with normal Pap test was 11.7% (95% CI 11.6%-11.7%), different from our findings (26.2%; 95% CI 19.2- 33.8).15-16 Our prevalence is similar to the prevalence observed by SubSaharan Africa (24.0%), and Eastern Europe (21.4%), but higher compared with the prevalence in Latin America (16.1%) [15,16]. In contrast to Gallegos-Bolaños, et al. our findings shown than the prevalence of HPV co-infection was lower than mono-infection and the most prevalent genotypes were HPV-6, and -16 [17]. In addition, the co-infection with HPV-16, and -61 genotypes were the most frequent combination contrary to reported by these authors, who indicated than the co-infection with HPV-51 and -52 genotypes was the most frequent combination in all their cases [17]. Similarly, Aziz H, et al. reported in a population of Punjab, Pakistan than HPV-6 was the most frequent HPV-genotype found in 25% of infected women, however, the prevalence observed in our population was lower (13.2%) [18]. Even more, HPV-16, -58, -59, and -31 genotypes were the most common HR-genotypes in patients with cervical lesions, while, types HPV-6, 61, and 53 were the most common LR-genotypes in the same group of patients (similar to a previously published study) [19]. Contrary to the results reported by Jácome Galarza, et al. [20]. cytological examination revealed normal Pap test in 36% of samples (35.7% with LSIL, and 28.3 % with HSIL), and a different double infections combination (to except of genotypes combination between HPV-53 and -62) [20]. In relation to triple infections, our findings were totally different from reported in the population of Michoacan [20]. Even, it was strange that the Cobas HPV test only identified 5 positives specimens among all samples. The detection rate of Cobas in our specimens is extremely low, however, we repeat all test (we check the reagents and procedure of our experiment). On the one hand, there are countries where the HPV co-infection is less frequent than the mono-infection, but there are regions where co-infection is higher [17, 21-22]. In Mexico the prevalence and patterns of HPV infection and cervical cancer distribution differ according to the geographic location analysed and determinants of health associated [17,19,23- 30]. It is well established than co-infection among HPV-genotypes is common in women, and men [17]. However, the knowledge of their epidemiological distribution is scarce. Some studies show that co-infection increases the risk of cervical cancer and is associated with both a low response and survival rate in those patients with cervical cancer that are receiving radiotherapy [17]. Our data in conjunction with other reports support evidence [17,27-28] to establish new appropriate prevention strategies for the design of new vaccines according to each population, as well as introduced new national cervical cancer screening programs and therefore restructure or design new national immunization programs. On the other hand, the present study provides epidemiological evidence indicating the need to include screening tests containing the most prevalent HPV-genotypes into national cervical cancer screening programs. As well as this study allowed established two pools of HR genotypes and two pools of LR genotypes, resulting in the screening of 25 different subtypes, which makes it more specific than any of the other tests available on the market. The pools were established according to the distribution of genotypes, clinical relevance, and technical procedure. In relation to genotype distribution, the main 10 genotypes were HPV-6, HPV-16, HPV-53, HPV-58, HPV-59, HPV-31, HPV-61, HPV-18, HPV-51, and HPV-56. Thus, one pool was established with the four most prevalent and relevant HR genotypes (16, 18, 31, and 59). Due to probes of HPV-58 and HPV-59 are similar, both were placed in different pool, and HPV-18 was included by its oncogenic relevance. The second pool of HR-genotypes was established for other 10 genotypes (33, 35, 39, 45, 51, 52, 56, 58, 66, and 68) based on their distribution. Likewise, the first pool of LR-genotypes was established with the main three genotypes (6, 53, and 61). Finally, the last pool of LR-genotypes includes eight genotypes (11, 40, 43, 44, 55, 70, 73, and 81) based on their distribution and clinical relevance (previous studies) [22]. As a result, is clear that using an HPV test that includes more genotypes will detect more cases of pre-cancerous cervical lesions, even than with traditional cytology. Similarly, is feasible to use this test at the population level making use of the infrastructure and resources already available within a cervical cancer screening program, and consequently the number of women referred for colposcopy owing to a positive result of HPV infection may be increased. According to the results reported by Koliopoulos, et al. [31]. in a systematic literature search of 40 studies with more than 140,000 women aged between 20 and 70 years old, for cervical intraepithelial neoplasias (CIN) of grade 2 or worse (CIN 2+) the pooled sensitivity for hybrid capture 2 (HC2), conventional cytology (CC) and liquid-based cytology (LBC) (atypical squamous cells of undetermined significance ASCUS+) were 89.9%, 62.5% and 72.9%, respectively, different to our findings (care GENETM-HPV test sensitivity 100% compared with COBAS system test) [31]. While pooled specificity estimates were 89.9%, 96.6%, and 90.3%, respectively [31]. Others authors report that the sensitivity of thin-layer Pap (with a result of ≥ASCUS) for identifying women with CIN 3 or higher was only 61.3% compared with 88.2% for HPV testing by PCR and 90.8% by signal amplification [32-33]. Furthermore, Mayrand, et al. [34,35] report that in women with HSIL, the sensitivity of HPV testing was 94.6%, whereas that of Pap testing was 55.4%.34 The specificity was 94.1% for HPV testing and 96.8% for Pap smears [34]. Compared with COBAS system test, the careGENETM-HPV test has a higher sensibility and a significantly more specific AUC. Immunization is the most cost-effective public health intervention. Nowadays, HPV vaccines in use around the world protect against infection from different genotypes: 16/18 (bivalent vaccine, Cervarix, GSK, Belgium), 6/11/16/18 (tetravalent vaccine, Gardasil, Merck, USA), and 6/11/16/18/31/33/45/52/58 (nonavalent vaccine, Gardasil 9, Merck, USA) [13-14, 35-36]. Present vaccination programmes against HPV do not include all prevalent genotypes of HPV, for instance, vaccines do not contain HPV-59 considered a HR-genotype. As a result, many patients infected with other HPV-genotypes do not benefit from current HPV vaccine programs. Even, the World Health Organization recommends HPV vaccines to be introduced into national immunization programs, where the cervical cancer prevention is a public health priority. Nonetheless, in order to meet this objective, it is necessary to have a broad coverage considering both, the age of the patients and the distribution of HPV genotypes. Our findings indicate that current vaccination programmes do not include several of the most prevalent types of HPV-genotypes, therefore, and unfortunately many adolescents and adults’ women fall outside the protection induced by vaccines. Consequently, our data show that it is more feasible to produce a vaccine with the prevalent genotypes in the population. Even thinking about producing the vaccine in the country based on the observed prevalence. In addition, for our population, follow-up studies will have to be developed to determine which genotype is the most oncogenic. In conclusion, we observed a different prevalence of HPV genotypes. Moreover, our study provides evidence that the “care GENETM- HPV detection-Kit-I test” for screening of human papillomavirus has more sensibility than other commercial tests such as COBAS system tests and was similar to Pap test. Consequently, this test can serve as a better triage test, because with this technique we can detect almost 90% of the HPV genotypes present in the population, and it is cheaper. Nevertheless, due to its low sensitivity compared to Pap test, it demands a strict follow-up of negative cases.
Acknowledgments
The authors would like to thank Professor Susana Ortiz Vela, Master in translation. Special thanks to Mr Young Choi and Dr Young Ho from Access bio and Wells Bio (Korea) and to Dr. Armando Ruiz Masieu from ISSSTE foundation.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Funding
The project was funded by “Fundacion ISSSTE A.C” (R.P.I 380.2019). The statistical analysis of this work was supported by “Centro de Investigacion y Educacion Continua S.C”, (CIE-2020001).
References
- Pan American Health Organization (2016) Integrating HPV testing in cervical cancer screening programs. A manual for program managers. Washington, D.C.,
- Burd EM (2003) Human papillomavirus and cervical cancer. Clin Microbiol Rev 16(1):1-17.
- Shikha Srivastava, U P Shahi, Arti Dibya, Sadhana Gupta, Jagat K Roy (2014) Distribution of HPV Genotypes and Involvement of Risk Factors in Cervical Lesions and Invasive Cervical Cancer: A Study in an Indian Population. Int J Mol Cell Med Spring 3(2):61-73.
- Cuschieri K, Wentzensen N (2008) Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol Biomarkers Prev 17(10): 2536-2545.
- Samuel Ratnam, Francois Coutlee, Dan Fontaine, James Bentley, Nicholas Escott, et al. (2011) Aptima HPV E6/E7 mRNA test is as sensitive as Hybrid Capture 2 Assay but more specific at detecting cervical precancer and cancer. J Clin Microbiol 49(2): 557-564.
- W Qu, G Jiang, Y Cruz, C J Chang, G Y Ho, R S Klein, et al. (1997) PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J Clin Microbiol 35(6): 1304-1310.
- Kyeong A So, In Ho Lee, Ki Heon Lee, Sung Ran Hong, Young Jun Kim, et al. (2019) Human papillomavirus genotype-specific risk in cervical carcinogenesis. J Gynecol Oncol 30(4): e52.
- Anco Molijn, Berhard Kleter, Wim Quint, Lee Jan van Doorn (2005) Molecular diagnosis of human papillomavirus (HPV) infections. J Clin Virology 32S: S43-S51.
- Bruni L, Albero G, Serrano B, et al. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in the World. Summary Report 17 June 2019. [April 3, 2021]
- López Hernández D, Beltrán Lagunes L, Brito Aranda L, López Hernández ML (2016) Human papillomavirus infection and its correlation with clinically relevant gynecological or obstetric situations: a cross-sectional study. Med Clin (Barc) 147(3): 101-108.
- Heredia-Caballero AG, Palacios-López GG, Castillo-Hernández MC, Hernández-Bueno AI, Medina-Arizmendi FV (2017) Prevalence and typing of human papillomavirus genotypes in women from the metropolitan area of the Valley of Mexico. Gyneco. obstet. Mex. [magazine on the Internet]. [cited Apr 032021]; 85(12): 809-818.
- Nubia Muñoz, F Xavier Bosch, Xavier Castellsagué, Mireia Díaz, Silvia de Sanjose, et al. (2004) Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer 111(2): 278-285.
- World Health Organization (2016) Meeting of the Strategic Advisory Group of Experts on Immunization, October 2016 conclusions and recommendations. Wkly Epidemiol Rec 48(2): 579-580.
- Comité Asesor de Vacunas (CAV-AEP) (2021) Virus del papiloma humano. Manual de vacunas en línea de la AEP [Internet]. Madrid: AEP; ene/2021. [consultado el abril 03, 2021].
- Laia Bruni, Mireia Diaz, Xavier Castellsagué, Elena Ferrer, F Xavier Bosch (2010) Cervical human papillomavirus prevalence in 5 continents: meta-analysis of 1 million women with normal cytological findings. J Infect Dis 202(12): 1789-1799.
- Silvia de Sanjosé, Mireia Diaz, Xavier Castellsagué, Gary Clifford, Laia Bruni, et al. (2007) Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: a meta-analysis. Lancet Infect Dis. 7(7): 453-439.
- Gallegos Bolaños J, Rivera Domínguez JA, Presno Bernal JM, Cervantes Villagrana RD (2017) High prevalence of co-infection between human papillomavirus (HPV) 51 and 52 in Mexican population. BMC Cancer 17:531.
- Hafsa Aziz, Huma Iqbal, Humera Mahmood, Shazia Fatima, Mohammad Faheem, et al. (2018) Human papillomavirus infection in females with normal cervical cytology: Genotyping and phylogenetic analysis among women in Punjab, Pakistan. Int J Infect Dis 66: 83-89.
- Israel, RG.S., Basilio, HS.J., del Rocío, TB.M. et al. (2020) A Pilot Study on the HPV Type Frequency in a Federal High-Specialty Hospital of Mexico City: Is HPV16 Our Main Problem?. SN Compr. Clin. Med. 2, 419-422.
- Irvin Jácome Galarza, María Ayumi Ito Nakashimada, Gloria Figueroa Aguilar, Ethel García-Latorre, Ma Isabel Salazar, et al. (2017) Prevalence of Human Papillomavirus in Women from the State of Michoacan, Mexico, Showed High Frequency of Unusual Virus Genotypes. Rev Invest Clin 69(5): 262-269.
- Adriana Aguilar Lemarroy, Verónica Vallejo Ruiz, Elva I Cortés-Gutiérrez, Manuel Eduardo Salgado Bernabé, Norma Patricia Ramos González, et al. (2015) Human papilomavirus infections in Mexican women with normal cytology, precancerous lesions, and cervical cancer: Type-specific prevalence and HPV coinfection. J Med Virol 87: 871-884.
- Elizabeth Louise Dickson, Rachel Isaksson Vogel, Robin L Bliss, Levi S Downs Jr (2013) Multiple-type human papillomavirus (HPV) infections: a cross-sectional analysis of the prevalence of specific types in 309,000 women referred for HPV testing at the time of cervical cytology. Int J Gynecol Cancer 23(7): 1295-1302.
- Rafael Gutiérrez Campos, Angélica Malacara Rosas, Elvia Gutiérrez Santillán, Mireya Delgado Gutiérrez, Rusland Enrique Torres Orozco, et al. (2019) Unusual prevalence of high-risk genotypes of human papillomavirus in a group of women with neoplastic lesions and cervical cancer from Central Mexico. PLoS One 14:(4): e0215222.
- Del R G LM, Rosado Lopez I, Valdez González N, Puerto Solís M (2004) High prevalence of human papillomavirus type 58 in Mexican colposcopy patients. J Clin Virol 29: 203-206.
- Navarro Vidal E, Hernandez Rosas F, Rey M, Flores Peredo L (2018) Prevalence of human papillomavirus genotypes in women from Cozumel, Mexico. Asian Pac J Cancer Prev 19: 2417-2422.
- C M Luna Aguirre, L M Reyes Cortés, A A Torres Grimaldo, S F Karr de León, R M Cerda Flores, et al. (2018) Prevalence of human papillomavirus types in north and central regions of Mexico. Epidemiol Infect 146: 1724-1730.
- López Rivera MG, Flores MOM, Villalba Magdaleno JD, Sánchez MV (2012) Prevalence of human papillomavirus in women from Mexico City. Infect Dis Obstet Gynecol 2012: 384758.
- Torroella Kouri M, Morsberger S, Carrillo A, et al. (1998) HPV prevalence among Mexican women with neoplastic and Normal cervixes. Gynecol Oncol 70: 115-120.
- López Hernández D (2013) Epidemiological association between body fat percentage and cervical cancer: a cross-sectional population-based survey from Mexico. Arch Med Res 44(6): 454-458.
- López Hernández D (2014) Type 2 Diabetes Mellitus and Habits Lifestyle Increases the Risk of Cervical Cancer: A Cross-Sectional Population-Based Study. Austin Journal of Obstetrics and Gynecology 1(3): 7.
- George Koliopoulos, Victoria N Nyaga, Nancy Santesso, Andrew Bryant, Pierre Pl Martin Hirsch, et al. (2017) Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst Rev 8(8): CD008587.
- Shalini L Kulasingam, James P Hughes, Nancy B Kiviat, Constance Mao, Noel S Weiss, et al. (2002) Evaluation of Human Papillomavirus Testing in Primary Screening for Cervical Abnormalities: Comparison of Sensitivity, Specificity, and Frequency of Referral. JAMA 288(14): 1749-1757.
- (2007) Specificity, sensitivity and cost. Nat Rev Cancer 7, 893.
- Marie Hélène Mayrand, Eliane Duarte-Franco, Isabel Rodrigues, Stephen D Walter, James Hanley, et al. (2007) Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N Engl J Med 357(16): 1579-1588.
- Nicolas Van de Velde, Marie Claude Boily, Mélanie Drolet, Eduardo L Franco, Marie-Hélène Mayrand, et al. (2012) Population-level impact of the bivalent, quadrivalent, and nonavalent human papillomavirus vaccines: a model-based analysis. J Natl Cancer Inst 104(22): 1712-1723.
- Mélanie Drolet, Jean François Laprise, Marie Claude Boily, Eduardo L Franco, Marc Brisson (2014) Potential cost-effectiveness of the nonavalent human papillomavirus (HPV) vaccine. Int J Cancer 134: 2264-2268.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.