Opinion 
 Creative Commons, CC-BY
Creative Commons, CC-BY
New SARS-Cov-2 Variant B.1.1.529: A Comparison with Previous Viral Variants Identified in The Apulia Region
*Corresponding author: Angelica Bianco, Antonio Parisi, Experimental Zooprophylactic Institute of Puglia and Basilicata, Via Manfredonia, Italy.
Received: November 29, 2021; Published: December 03, 2021
DOI: 10.34297/AJBSR.2021.15.002062
Opinion
Following the epidemic COVID-19 the emergence of multiple variants has been reported. Mutations of the virus may cause changes in its infectivity and pathogenicity, resulting in the emergence of highly infectious or lethal mutant strains, they may also change the antigenicity of the virus, leading to failures of existing antibody treatments or the vaccine [1-3]. There are several mutations of SARS-CoV-2 genome that have received attention from scientists worldwide. The first mutation of interest was D614G in the Spike protein [4]; strains that harboured this mutation became the dominant strain globally with a prevalence >95% [3]. Successively, a growing interest has been attributed to nucleotide variants identified in the Spike gene because these variants may influence the efficacy of vaccines and therapeutic monoclonal antibodies [5,6]. Thus, additional mutations of concern were identified and linked to several Variants of Concern (VOCs) [7] including Alpha (B.1.1.7).
First identified in the UK [8] but found more than 50 countries, Beta (B.1.351) [9]. First identified in South Africa, but also found at least 20 countries, and Gamma (P.1) [10]. First identified in Brazil but, similarly to the others, has spread to multiple countries. The last emergent variant of SARS-CoV-2 is represented by the VOC Omicron variant (Lineage B.1.1.529) identified in Botswana in Sud-Africa [11] and defined as “variant of horror”, because it seems to spread much faster than the previous ones. This VOC is characterized by 32 mutations to the spike protein (Table 1). Many of the changes have been previously found in variants such as Delta and Alpha and are related to increased infectivity and ability to evade infection-blocking antibodies. Interestingly, the attention has been focused on two mutations in S1/S2 regions of the gene in amino acid positions N679K and P681H.
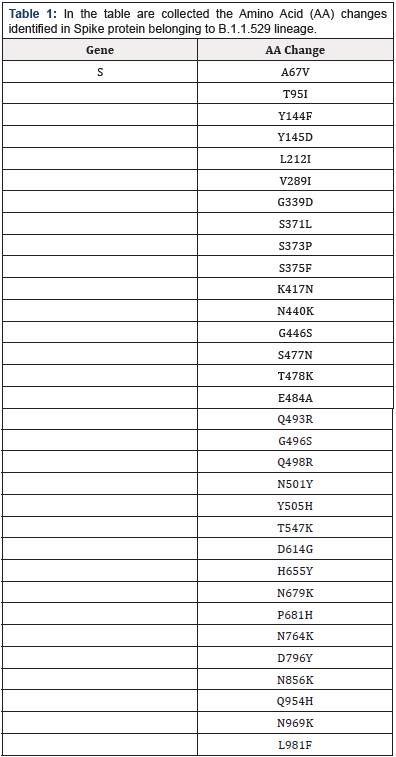
Table 1. In the table are collected the Amino Acid (AA) changes identified in Spike protein belonging to B.1.1.529 lineage.
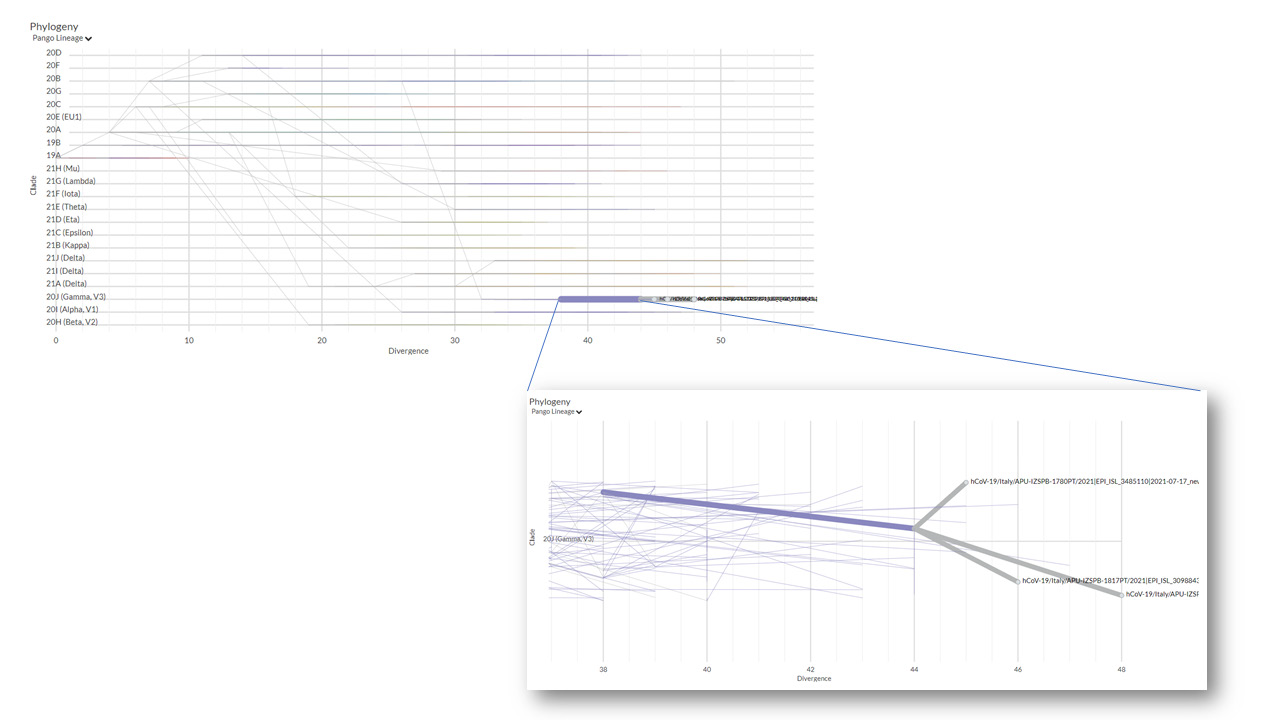
Our laboratory is involved in genomic surveillance of circulating SARS-CoV-2 in Apulia region in Italy [12]. To date we have sequenced more than 1500 SARS-CoV-2 genomes labelled with “APU-IZSPB” (https://www.gisaid.org/). We surveyed Spike mutations N679K and P681H in our genomes. We identified the mutation P681H in 516 genomes belonging to B.1.1.7(99%), B.1(0.6%) and P.1.1(0.4%) lineages, according to the occurrence of this mutation in different variant of SARS-CoV-2 as reported in https://outbreak. info/. Interestingly, we detected the N679K mutation in only three genomes (EPI_ISL_3485110; EPI_ISL_3098848; EPI_ISL_3098843) belonging to P.1.12 lineage (Figure 1) sequenced in July 2021. This funding is surprising since this mutation is not common in the mentioned lineage (https://outbreak.info/situationreports? pango=P.1).
Furthermore, the P1+N679K lineage does not seem to have spread in our Region as it is no longer identified in subsequent surveillance plans. However, this suggests that new mutations may occur and may cause some variants to be more virulent. Additionally, the combination of two or more virulent mutations may cause the emergence of more infectious genotypes. For this reason, gathering and sharing information on genome variants and on S gene will provide important data on the emergence of possible new variants and subvariants. Genomic surveillance will allow the evaluation the effectiveness of vaccines and the monitoring of the changes related to the pathogenesis of the disease. Furthermore, it will prove useful to determine the rate and speed of mutations emergences since they play an important role in the virus escaping host immune response and thus developing resistance to drugs.
Conflict of interest
None
Acknowledgement
None
References
- Garcia Beltran WF, Lam EC, Denis K, Nitido AD, Garcia ZH, et al. (2021) Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184(9): 2372-2383.
- Gianmarco L, Maria OM, Annarita B, Guido A, Alessandro B, et al. (2020) The impact of the SARS-CoV-2 pandemic on healthcare provision in Italy to non-COVID patients a systematic review. Med Rxiv.
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, et al. (2020) Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182(4): 812-827.
- Zhang L, Jackson CB, Mou H, Ojha A, Rangarajan ES, et al. (2020) The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. Bio Rxiv.
- Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, et al. (2021) SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19(7): 409-424.
- Ahmad L (2021) Implication of SARS-CoV-2 Immune Escape Spike Variants on Secondary and Vaccine Breakthrough Infections. Front Immunol 12: 742167.
- Mattiuzzi C, Henry BM, Lippi G (2021) Is diffusion of SARS-CoV-2 variants of concern associated with different symptoms? J Infect.
- Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, et al. (2021) Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep 70(3): 95-99.
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, et al. (2021) Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592(7854): 438-443.
- Faria NR, Mellan TA, Whittaker C, Claro IM, Candido D from S, et al. (2021) Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 372(6544): 815-821.
- Callaway E (2021) Heavily mutated omicorn variant puts scientists on alert. Nature.
- Capozzi L, Bianco A, Del Sambro L, Simone D, Lippolis A, et al. (2021) Genomic Surveillance of Circulating SARS-CoV-2 in Southeast Italy a One-Year Retrospective Genetic Study. Viruses 13(5): 731.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.