Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Tumor Necrosis Factor Affects Steroidogenic Factor-1 Expression in Ovarian Granular Cells Via Inducing Gene Methylation
*Corresponding author: Xiaoxi Sun, Affiliated Obstetrics and Gynecology Hospital of Fudan University, Shanghai JIAI Genetics and IVF institute, Shanghai, People’s Republic of China.
Received: February 28, 2022; Published: March 11, 2022
DOI: 10.34297/AJBSR.2022.15.002154
Abstract
Objective: The expression of SF-1 in ovarian granulosa cells is critical for follicular growth and development, and the decreased expression of SF-1 in ovarian granulosa cells under endometriosis (EMs) pathology is not clear. This study aims to investigate the mechanism of tumor necrosis factor (TNF), a factor promoting endometriosis, in regulating the promoter methylation and expression of steroidogenic factor-1 (SF-1) in ovarian granulosa cells.
Methods: Human ovarian granulosa cell line KGN was used. Methylation inhibition was mediated by cells treated with 5-Aza-2 ‘deoxycytidine (5-Aza). The inhibition of the p65 expression gene in KGN cells was mediated by a lentiviral CRISPR/Cas9 vector (p65- /-KGN cells). The methylation level in the promoter region of SF-1 was detected by BSP assay and SF-1 expression was determined using quantitative real-time PCR and western blot.
Results: In KGN cells, inhibition methylation increases the mRNA level of SF-1. TNF treatment causes an increased methylation level of SF-1 and inhibits the mRNA and protein expression of SF-1 in a concentration-dependent manner. Inhibition methylation of wild-type KGN (p65wt) and p65-/- KGN cells enhances SF-1 expression. TNF treatment significantly increase the methylation level of SF-1 and decreased the expression of SF-1 in p65wt KGN cells but does not affect that in p65-/- KGN cells.
Conclusion: TNF induces the increase in methylation of the SF-1 gene, which leads to the decrease of SF-1 expression in KGN cells through the ReIA/p65 pathway. It is suggested that the decrease of SF-1 expression in ovarian granulosa cells of EMs is related to the immune mechanism of EMs disease.
Keywords: Endometriosis; Ovarian granulosa cells; Steroidogenic factor 1; Tumor necrosis factor
Introduction
Endometriosis (EMs) is an immune-related chronic inflammatory disease and can lead to infertility in reproductiveage women through multiple factors and mechanisms [1]. In vitro fertilization embryo transfer (IVF-ET) is one of the effective treatment methods for EMs-induced infertility, but EMs has adverse effects on the treatment outcome of IVF-ET. The research has pointed out that EM-derived egg embryos had a limited growth rate and clinical pregnancy rate in recipients with non-EMs, mostly because of follicular development obstacles and decline in the quality of oocytes leading to a decline in embryonic plant capacity [2,3]. This suggests the necessity of conducting basic research to explore the mechanism of egg quality loss in EMs-induced infertility.
Steroidogenic factor-1 (SF-1), a member of the myonuclear receptor family, is abnormally expressed in intrinsic and ectopic endometrial tissue of EMs [4] and plays an important role in the pathogenesis of EMs [5]. In the ectopic endometrium, SF-1 promotes the high expression of its target gene P450 aromatase, which enables the ectopic endometrium to synthesize estrogen locally without relying on circulating estrogen, and further promotes the growth of the ectopic endometrium [5]. Our previous study found that the transcription level of SF-1 in ovarian granulosa cells of patients with EMs was downregulated and the level of estradiol synthesis in granulosa cells was decreased [6,7]. The significance of SF-1 expression in ovarian granulosa cells is its necessity for normal follicular development and the maintenance of ovarian reproductive function, which is different from its promoting role in the pathological growth of the endometrium. SF-1 knockout mice often show follicular arrest, follicular atresia, and infertility [8].
Herein, understanding the molecular mechanism of the decreased expression of SF-1 in ovarian granulosa cells in the pathological state of EMs can help us to understand the pathological mechanism of infertility induced by EMs. The regulation of SF-1 gene expression is mainly realized through DNA methylation. As one of the important epigenetic mechanisms, the methylation of CpG island in gene promoters is an important way to regulate gene expression. A study has shown that the CpG site in the promoter region of the SF-1 gene is hypermethylated [9]. Whether the hypermethylated state of the SF-1 promoter is related to the decreased expression of SF-1 and the underlying mechanisms of SF-1 expression in granulosa cells of EMs population has not been known yet.
Existing theories support that the inflammatory response involved by macrophages plays an important role in the occurrence and development of EMs [10,11]. Tumor necrosis factor (TNF), a cytokine secreted by macrophages, is an on-off activating factor in early inflammation. The level of TNF in follicular fluid of EMs was significantly higher than that in normal follicular fluid [12,13] which is involved in the pathogenesis of EMs and is one of the initiating factors for the occurrence, development, and invasion of ectopic endometrium. TNF has a function in regulating the expression of its target genes by activating transcription factors (i.e., nuclear factor-kappa B, NF-κB) to change the methylation level of target gene promoters [14,15]. In this study, we tried to investigate whether TNF affected the methylation level of the SF-1 gene promoter, thereby regulating the transcription and expression of SF-1 in granulosa cells.
Material and Methods
Reagents
Considering the obtained ovarian granulosa cells purified from follicular fluid in IVF puncture egg extraction is luteinated granulosa cells and may have an influence on the accuracy and significance for the experiment, so the selection of ovarian granular cell lines (KGN) as the research object. KGN cells were donated by Professor Yi-ming Mu of the Chinese people’s liberation army general hospital. Cell transfection reagent Lipofectamine2000, DMEM high glucose medium, fetal bovine serum, trypsin, and PBS buffer was purchased from Thermo Fisher Scientific (USA). P65 lentivirus kit was provided by Shanghai Heyuan Biotech (China). ECL substrate chromophase solution and Trizol kit were purchased from Sigma (USA). PCR reagent and reverse transcription kit were purchased from Takara (China). TIAN amp Genomic DNA Kit was purchased from Beijing Tiangen Biochemical Technology (China). EZ DNA Methylation Gold kit was purchased from Zymo Research, Inc (USA).
Cell Culture and Drug Treatment
About 104 KGN cells were resuspended in a DMEM culture medium (containing 10% FBS and 1% double-antibody) and cultured in an incubator at 37℃ and 5% CO2. The medium was changed every 2-3 days. When the cells grew to about 90% confluence, the cells were passed or frozen stored. Before the cell intervention experiment, the cells were starved with the DMEM culture medium for 12-24h.
To explore the effect of inhibition of methylation on KGN cell expression of SF-1, cells were treated with 1μM 5-Aza-2 ‘deoxycytidine (a DNA methylation inhibitor, 5-aza). Cells treated with an equal volume of DMSO served as the negative control.
Cell Transfection for SF-1 Knockdown
A total of 5×104 KGN cells was inoculated on a 24-well plate one day before transfection, and 50, 100, and 200 nmol/L siRNA (siRNA for SF-1 designed by Suzhou Jima Company, China) were added to 50μL DMEM serum-free medium. Lipofectamin2000 reagent was diluted with serum-free DMEM (1μL:50μL) (2.4μg RNAi-mate reagent was added when DNA transfection). The diluted siRNA and lipofectamin2000 reagent were mixed to form the siRNA/ lipofectamine (or DNA/ lipofectamine) complex, and the complex was added to the wells containing the cells and the culture medium.
Lentiviral CRISPR/Cas9 Mediated p65 Inhibition
Lentiviral CRISPR/Cas9 vector was used to inhibit p65 in KGN cells (p65-/- KGN cells) with empty vector in KGN cells (p65wild type(wt.) KGN cells) as control. In brief, a total of 3~5×104 KGN cells in 500μL medium were inoculated in a 24-well culture plate. Lentivirus infection begins when cells grew to 30-50% confluence. KGN cells were transfected with p65 CRISPR/Cas9 lentivirus or control vector according to the instructions of the lentivirus kit. After mixing with vectors, the cells were placed in an incubator (37℃, 5% CO2) overnight and the culture medium was changed 12-20 hours later. The infection effect was observed 24 hours later by laser confocal microscopy and GFP fluorescence.
Quantitative Real-Time PCR Detects SF-1 Transcription
Total RNA was extracted by the Trizol method, and cDNA was synthesized referring to the instructions of the TAKARA reverse transcription kit (RR047A, TaKaRa, China). SF-1 primer was synthesized by Suzhou Jinweizi Company (China). SF-1 Forward: 5’-TGGACHHAATCGGAACACG-3’; SF-1 Reverse: 5’ TGGCTATGGCACCTTGAAAAAC-3’. the reaction conditions of general quantitative PCR reagent system were as follows: pre-denaturation at 95℃ for 10 minutes, denaturation at 95℃ for 25 seconds, annealing at 55℃ for 25 seconds, extension at 72℃ for 30 seconds, circulation for 40 times, extension at 72℃ for 5 minutes to end the reaction, and fluorescence was collected at this step. GAPDH gene was an internal reference.
Western Blot Detects SF-1 Protein Content
KGN cells were transfected with SF-1 siRNA plasmid and was added to with cell lysate to extract total proteins. The protein sample was quantified by the BCA method and a volume of 40μL was loaded on PAGE gel. After electrophoresis, proteins were transferred to the PVDF membrane (Merck, German). Primary antibodies against SF-1 and GAPDH (Proteintech, USA) were added to the membrane and incubated at 4℃ overnight. On the next day, the PVDF membrane was incubated with a secondary antibody (HRP-labeled sheep anti-rabbit IgG) (Biotin, China) for 2 hours. PVDF membrane was washed at room temperature and added with ECL. The blot was visualized and photographed.
Bisulfite Sequencing PCR (BSP) Detects Methylation Level at 5 ‘CpG Site of SF-1 Gene
The centrifugal type of column genomic DNA extraction kit was used for the extraction of genomic DNA, and the EZ DNA Methylation-Gold kit for bisulfite conversion. The genomic DNA after bisulfite conversion was used for specific PCR amplification (SF-1 primer and the PCR reaction conditions were the same as the former). PCR products were purified and connected to PMD18-T carrier. The positive plaque was selected for plasmid extraction and sequencing (10 cloning).
Statistical Analysis
Data were analyzed on SPSS version 11.0 and presented as mean standard deviation (SD). Analysis of significant differences between groups was conducted using a student’s t-test. More than 3 independent experiments were performed for each protocol. A p-value less than 0.05 was considered statistical significance.
Results
Methylation Inhibition Promotes SF-1 Expression
We examined the SF-1 expression levels in methylation inhibitor 5-aza treated cells. The results showed that compared with DMSO, 5-aza induced the increase in mRNA and protein levels of SF-1 (Figure 1a and 1b, P<0.01), suggesting that hypomethylation promoted SF-1 transcription and protein expression in KGN cells.
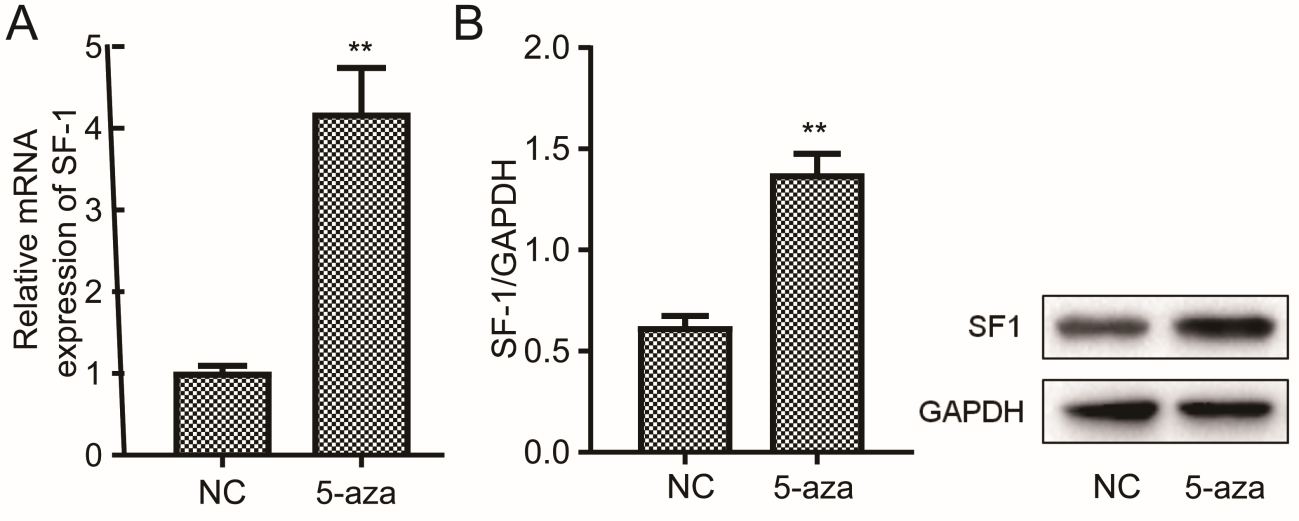
Figure 1: Effect of methylation inhibitor on SF-1 expression. KGN cells were treated with 1 μM of 5-aza-2’deoxycytidine and that treated with DMSO as a negative control (NC). Cells were collected for analysis of expression level of mRNA (A) and protein (B) using qRT-PCR and western blot. **P<0.01.
TNF Enhances Methylation Level of the SF-1 Gene
BSP assay detected the effect of different concentrations of TNF on methylation level of the proximal promoter region of SF-1 gene via sequencing on gene fragments covering 13 CpG loci (from CpG-84 to the CpG+168) near transcription start site and part of the exon 1 area (122-/129/+). The result showed that 10ng/mL, 20ng/mL, 30ng/mL, and 40ng/mL TNF caused the increase in the methylation level of the SF-1 gene and the methylation level on the SF-1 gene was increased with the increase of TNF concentration in KGN cells (Figure 2).
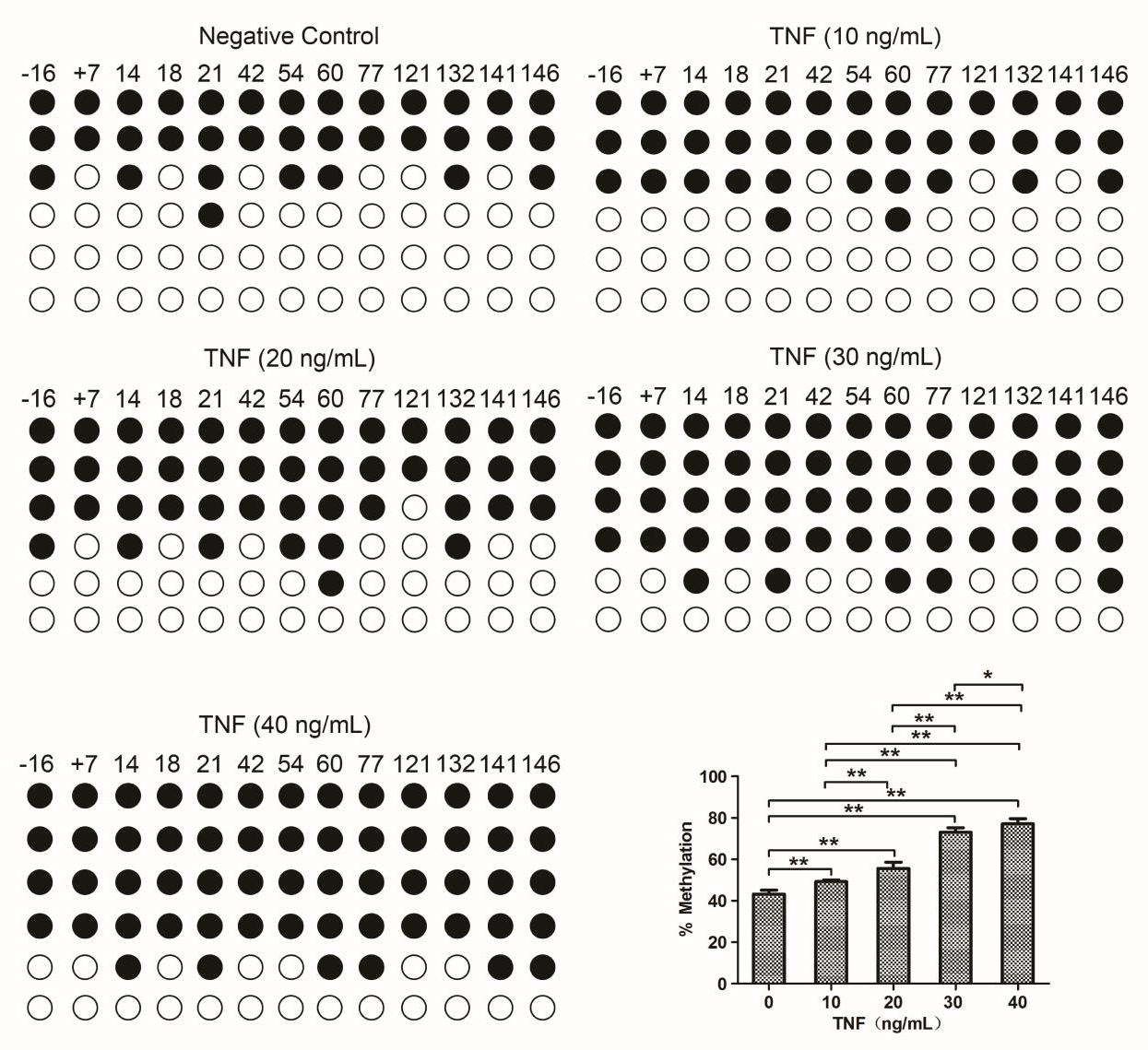
Figure 2: Effects of different concentrations of TNF on SF-1 methylation in KGN cells. Cells were treated with TNF as indicated doses and that treated with DMSO as a negative control (NC). Cells were collected for a measure of methylation level at 5’CpG site of SF-1 gene using BSP assay. *P<0.05, **P<0.01.
TNF Inhibits SF-1 Expression
Quantitative real-time PCR and Western blot determined SF-1 expression in KGN cell processing different concentrations (10 ng/ ml, 20 ng/ml, 30 ng/ml, and 40 ng/ml) of TNF. The results showed that compared with negative control, cells in the TNF treatment group expressed a lower level of SF-1 mRNA and protein. The inhibitory effect of TNF on SF-1 mRNA and protein expression was in a dose-dependent manner and the inhibition effect was strongest at 30ng/ml (Figure 3A and B).
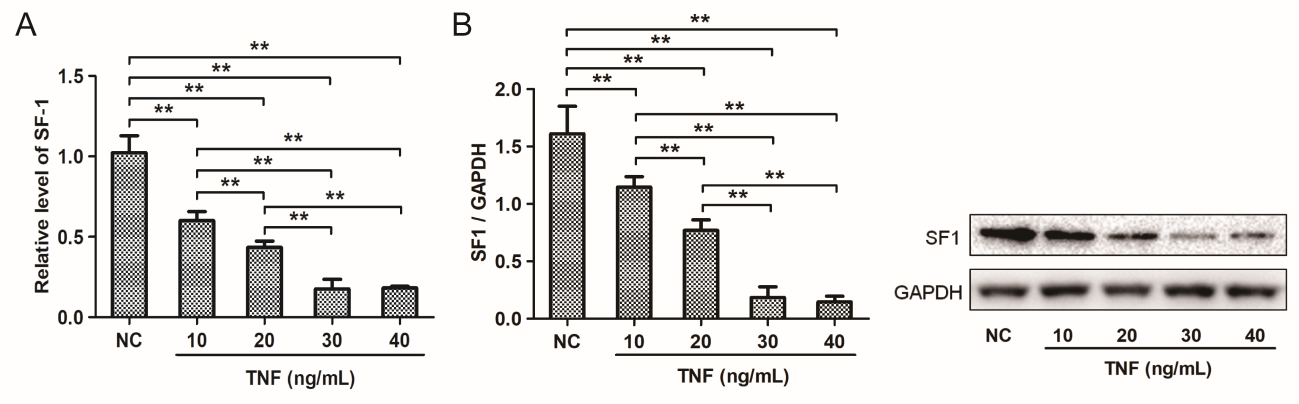
Figure 3: Effects of different concentrations of TNF on SF-1 expression in KGN cells. KGN cells were treated with TNF as indicated doses and that treated with DMSO as the negative control (NC). Cells were collected for analysis of expression level of mRNA (A) and protein (B) using qRT-PCR and western blot. **P<0.01.
TNF Affects Methylation Level and SF-1 Expression in p65wt And p65-/- KGN Cells

Figure 4: TNF affects methylation level and SF-1 expression in p65wt and p65-/- KGN cells. (A)qRT-PCR detects SF-1 mRNA expression in cells treated with methylation inhibitor 5-aza-2’deoxycytidine. **P<0.01. (B) BSP assay examines methylation levels in TNF-treated cells. *P<0.05; **P<0.01. (C) qRT-PCR detects SF-1 mRNA expression in TNF-treated cells. **P<0.01.
The above results suggest that TNF may inhibit the transcription and expression of SF-1 in KGN cells by regulating the methylation level of the SF-1 promoter. Studies have shown that NF-κB (a transcription factor with an active subunit of REIA/ p65) was cascaded by TNF and thereby promoted the expression of its target genes [14,15]. Our results showed that p65-/-KGN cells expressed higher levels of SF-1 than p65wt cells when the negative control treatment and after methylation inhibitor 5-aza treatment, SF-1 expression levels were significantly increased in both of p65wt and p65-/-KGN cells. These data suggest that p65 plays a role in the methylation and expression of SF-1 and the regulatory expression pathway by p65 is still effective even in p65-/-KGN cells (Figure 4a).
To confirm whether REIA/p65 is involved in the process of TNF induced the increase in SF-1 methylation and the decrease in SF-1 expression in KGN cells, we treated p65wt and p65-/-KGN cells with different concentrations of TNF (10ng/mL, 20ng/mL, 30ng/ mL, and 40ng/mL) to detect the methylation level of SF-1. The result of the BSP assay showed that in p65wt KGN cells, 10ng/mL of TNF did not change the methylation level of SF-1, while 20, 30, and 40ng/mL of TNF significantly increased the methylation level of SF-1 (Figure 4b, P<0.01). However, in p65-/- KGN cells, TNF did not affect SF-1 methylation and SF-1 expression (Figure 4b and 4c). Besides, at any concentration of TNF, the SF-1 methylation level of p65wtKGN cells was significantly higher than that of p65-/- KGN cells, while the SF-1 expression level of p65wtKGN cells was lower than that of p65-/- KGN cells (Figure 4b, 4c, P<0.01). It is suggested that p65 is an important switch in TNF regulation of SF-1 methylation and expression in KGN cells.
Discussion
Inflammation and immune cell dysregulation affect infertility associated with EMs. The level of cytokine TNF is significantly increased in the peritoneal fluid of patients with EMs complicated with infertility and is closely related to the occurrence of EMs [16,17]. The results in this study show that TNF decreases the expression of SF-1, an essential molecular for follicular development, via increasing the methylation level of the SF-1 gene in KGN cells suggesting that TNF-induced downregulation of SF-1 may be a pathological mechanism of EMs-induced infertility.
Previously, we detected high methylation status in the CpG sites of SF-1 gene promoter region in the ovarian granular cells of EMs patients using the BSP cloning sequencing method, especially focused in CpG loci near the transcription start site +77, +121, and +141 [18]. In this study, the low methylation level of SF-1 promotes SF-1 mRNA and protein expression in KGN cells suggesting that gene methylation level affects SF-1 mRNA and protein expression. This result is in accord with the finding by Xue et al., in which the SF-1 gene is demethylated in the interstitial cells of the intrinsic and ectopic endometrium, and this abnormal demethylated state is an important mechanism for the expression of SF-1 in ectopic endometrium (while the hypermethylated SF-1 gene in normal endometrium silences the expression of SF-1) [9,19].
We found that TNF can significantly reduce the mRNA and protein expression level of SF-1 in KGN cells. In the pathogenesis of EMs, TNF is one of the initiating factors for the occurrence, development, and invasion of ectopic endometrium. In the follicular fluid of EMs patients, the level of TNF is significantly increased [12, 13]. Our pre-test supported this fact (TNF 363±87ng/L of EMs vs. 78±36ng/L of normal, P<0.05). As a switch-acting promoter for immune response, TNF forms a cascade reaction with transcription factor NF-κB via activating its active subunit REIA/p65, leading to abnormal expression of the target genes [14, 15]. The cascade reaction induced by TNF and NF-κB-mediated transcription inhibition of targeted genes is commonly found in tumors [15,20]. By searching the bioinformatics software, we found that NF-κB has binding sites in the SF-1 gene near the transcription initiation site (search at www.genecards.org). This leads us to further investigate whether TNF associated with SF-1 methylation also occurred in ovarian granulosa cells.
Campbell et al., for the first time put forward the definition of REIA/p65 to “active repressor [21], and study in tumor cells found that REIA/p65 signal pathway plays an important role in cell proliferation, differentiation, apoptosis under the stimulation of cytokines (i.e., TNF and IL- 6) [14]. The molecular mechanism by which REIA/p65 acts is to suppress the transcription level of the gene by enhancing the methylation of the gene promoter. In KGN cells, we found that TNF increased the methylation level of the SF-1 gene in a concentration-dependent manner indicating that TNF inhibits the transcription and expression of SF-1 by controlling SF-1 gene methylation levels. REIA/p65 is a key molecule in this process. TNF did not affect the methylation level and expression of the SF-1 gene in p65-/- KGN cells. The effect of TNF on enhancing the methylation level of the SF-1 gene and decreasing the expression of SF-1 occurred only in p65wt KGN cells, although the methylation of p65-/- KGN cells was still effective in regulating the expression of SF-1. It is suggested that the REIA/p65 pathway plays a key role in the increase of SF-1 methylation and the downregulation of SF-1 expression in KGN cells induced by TNF, which is consistent with the issue that REIA/p65 is a potential regulatory factor of DNA methylation in oncologic studies.
In summary, the present research found that TNF can induce an increase in gene methylation and thereby contributes the decrease in SF-1 mRNA and protein expression levels via REIA/p65 pathway in KGN cells suggesting that the reduced expression of SF-1 in ovarian granulosa cells is related to the inflammation and immune cell dysregulation pathomechanism of EMs disease. Whether drugs can be used to reduce the TNF level of follicular fluid of EMs and restore the normal expression of SF-1 in granulosa cells, to improve the quality of eggs and embryos is a research direction worthy of exploration.
Acknowledgement
None
References
- Y Wang, K Nicholes, I M Shih (2020) The Origin and Pathogenesis of Endometriosis. Annu Rev Pathol 15: 71-95.
- E E Hauzman, J A Garcia Velasco, A Pellicer (2013) Oocyte donation and endometriosis: What are the lessons? Semin Reprod Med 31(2): 173-177.
- S Senapati, M D Sammel, C Morse, K T Barnhart (2016) Impact of endometriosis on in vitro fertilization outcomes: an evaluation of the Society for Assisted Reproductive Technologies Database. Fertil Steril 106(1): 164-171 e1.
- Y Tian, B Kong, W Zhu, S Su, Y Kan (2009) Expression of steroidogenic factor 1 (SF-1) and steroidogenic acute regulatory protein (StAR) in endometriosis is associated with endometriosis severity, J Int Med Res 37(5): 1389-1395.
- S E Bulun, H Utsunomiya, Z Lin, P Yin, Y H Cheng, et al. (2009) Steroidogenic factor-1 and endometriosis. Mol Cell Endocrinol 300(1-2): 104-108.
- X Lu, Y Wu, X H Gao, Y W Wang, L Wang, et al. (2012) Effect of letrozole on estradiol production and P450 aromatase messenger RNA expression of cultured luteinized granulosa cells from women with and without endometriosis. Fertil Steril 98(1): 131-135.
- X Lu, Z M Wu, Y W Wang, M Wang, W W Cheng, et al. (2015) Liver receptor homologue-1 and steroidogenic factor-1 expression in cultured granulosa cells from patients with endometriosis: A preliminary study. J Obstet Gynaecol Res 41(12): 1927-1934.
- M C Meinsohn, O E Smith, K Bertolin, B D Murphy (2019) The Orphan Nuclear Receptors Steroidogenic Factor-1 and Liver Receptor Homolog-1: Structure, Regulation, and Essential Roles in Mammalian Reproduction. Physiol Rev 99(2): 1249-1279.
- Q Xue, Y Xu, H Yang, L Zhang, J Shang, et al. (2014) Methylation of a novel CpG island of intron 1 is associated with steroidogenic factor 1 expression in endometriotic stromal cells. Reprod Sci 21(3): 395-400.
- C Hogg, A W Horne, E Greaves (2020) Endometriosis-Associated Macrophages: Origin, Phenotype, and Function. Front Endocrinol (Lausanne) 11: 7.
- J Vallve Juanico, X Santamaria, K C Vo, S Houshdaran, L C Giudice (2019) Macrophages display proinflammatory phenotypes in the eutopic endometrium of women with endometriosis with relevance to an infectious etiology of the disease. Fertil Steril 112(6): 1118-1128.
- H Falconer, J Sundqvist, K Gemzell Danielsson, B von Schoultz, T M D Hooghe, et al. (2009) outcome in women with endometriosis in relation to tumour necrosis factor and anti-Mullerian hormone. Reprod Biomed Online 18(4): 582-588.
- A K Singh, M Dutta, R Chattopadhyay, B Chakravarty, K Chaudhury, et al. (2016) Intrafollicular interleukin-8, interleukin-12, and adrenomedullin are the promising prognostic markers of oocyte and embryo quality in women with endometriosis. J Assist Reprod Genet 33(10): 1363-1372.
- L Xia, S Tan, Y Zhou, J Lin, H Wang, et al. (2018) Role of the NFkappaB-signaling pathway in cancer. Onco Targets Ther 11: 2063-2073.
- Y Liu, M W Mayo, A S Nagji, P W Smith, C S Ramsey, et al. (2012) Phosphorylation of RelA/p65 promotes DNMT-1 recruitment to chromatin and represses transcription of the tumor metastasis suppressor gene BRMS1. Oncogene 31(9): 1143-1154.
- X M Wang, Z Y Ma, N Song, (2018) Inflammatory cytokines IL-6, IL-10, IL-13, TNF-alpha and peritoneal fluid flora were associated with infertility in patients with endometriosis. Eur Rev Med Pharmacol Sci 22(9): 2513-2518.
- B Babaabasi, A Ahani, F Sadeghi, H Bashizade Fakhar, (2019) The Association between TNF-alpha Gene Polymorphisms and Endometriosis in An Iranian Population. Int J Fertil Steril 13(1): 6-11.
- X Lu, Wu Z M, Wang W, Cheng W W, (2016) Study on the methylation status of SF-1 gene promoter in ovarian granulosa cells in patients with endometriosis. Reproduction Contraception 36(6): 812.
- Q Xue, Z Lin, P Yin, M P Milad, Y H Cheng, et al. (2007) Transcriptional activation of steroidogenic factor-1 by hypomethylation of the 5' CpG island in endometriosis, J Clin Endocrinol Metab 92(8): 3261-3267.
- H Wehbe, R Henson, F Meng, J Mize Berge (2006) T Patel, Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res 66(21): 10517-10524.
- K J Campbell, S Rocha (2004) N D Perkins Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol Cell 13(6): 853-865.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.