Mini Review 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Klebsiella Pneumoniae Infections: A Future Threat to Global Health?
*Corresponding author: Ana Paula Mattos Arêas, Center for Natural and Human Sciences-Federal University of ABC (UFABC), Brazil.
Received:June 14, 2022; Published: July 08, 2022
DOI: 10.34297/AJBSR.2022.16.002273
Abstract
Klebsiella pneumoniae infections constitute a great concern, especially in developing countries. Multidrug-resistant strains that possess a formidable ability to exchange DNA and high genomic diversity are particularly challenging to clinical practitioners and researchers to face and overcome. In this study, we discuss relevant aspects of aetiology and initiatives to control the dissemination of this pathogen. Given the burden of nosocomial and community-acquired K. pneumoniae infections as well as the capacity to mune other bacteria with resistant cassettes such as carbapenem, it seems adequate to affirm that it is a present threat to global health, since K. pneumoniae is no longer an emergent pathogen.
Keywords: Klebsiella Pneumoniae; Bloodstream Infections; Vaccines; Antibiotics, Global Health; Nosocomial; Carbapenem; Bacterium; ESKAPE Species; Antimicrobial Molecules; Genomic; Clinical Manifestations
Introduction
Klebsiella pneumoniae is an enteric capsulated Gram-negative bacterium that is a major supergerm. Traditionally recognized as a relevant pathogen in hospital environment, K. pneumoniae strains-mainly the multidrug-resistant ones-are currently related to mild and severe infections in the community. Pediatrics invasive diseases are especially of great concern [1]. Nosocomial and community-acquired K. pneumoniae infections, frequently initiated in the oropharynx or intestinal niche, include device-associated contamination, urinary tract diseases, pneumonia, bloodstream infections, and other clinical manifestations. A population surveillance-based study showed that patients with co-morbidities tend to be more susceptible to bloodstream infections, caused by Klebsiella species [2]. Several antibiotic resistant K. pneumoniae strains have been circulating, which constitutes a challenge to treat pathological alterations of patient’s tissues infected with one or more ESKAPE species (reviewed in [3]). ESKAPE is an acronym for some species of pathogenic drug-resistant hospital-circulating bacteria, which are the quintessence of transmission, acquisition of resistance and pathogenesis adaptation: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species [4]. ESKAPE bacteria utilize a plethora of molecular mechanisms to avoid the action of antimicrobial molecules, including structure alteration, inactivation or change in their capacity to bind the molecular targets; lesser permissiveness to accumulate drugs and higher ability to export them; biofilm formation etc., [5].
Burden of K. Pneumoniae Infections
ESKAPE bacteria are involved in efficient horizontal gene transfer processes, which are responsible for the spread of resistance cassettes. These sequences could potentially confer life- threatening characteristics to less pathogenic resident bacteria. According to PAHO (2021) [6], misuse of antibiotics, during SARSCoV2 pandemics, also contributed to this scenario of disseminated infections caused by multidrug-resistant bacterial species. In this sense, WHO (2019a) [7] estimates that antimicrobial resistance, found not only in bacteria, but also in virus, fungi, and parasites, were amongst the top ten threats to global health in 2019. A similar report, prepared by CDC (2019) [8], stated carbapenemresistant Enterobacteriaceae-including K. pneumoniae strains-as an urgent threat. Certainly, a post-pandemic period will not portrait a distinct landscape. Therefore, given the scenario imposed by ESKAPE infections, it is essential to monitor and control the use of antimicrobial drugs in humans and animals as well. Several initiatives-with a wide-ranging effectivity-have been described to treat ESKAPE infections, from antibiotics of varied spectra of actions to photodynamic light therapy [9].
Although these treatments offer alternative ways to deal with this issue, vulnerable population, such as the oldest and youngest groups besides immunocompromised individuals, remains to be more prone to get infected and to develop severe symptoms. According to WHO (2019b) [10], neonatal conditions were the 5th global cause of deaths due to communicable diseases in 2019. In low-income countries, these illnesses were the leading cause of casualties. K. pneumoniae is a relevant causative agent in this scenario. A cohort study, performed in 12 sites of seven African and Asian developing countries, analysed the incidence of neonatal sepsis caused by some bacterial pathogens [11]. The Burden of Antibiotic Resistance in Neonates from Developing Societies (BARNARDS) project showed that K. pneumoniae was the most isolated Gram-negative species in the sample. Astonishingly, data showed that all bacterial isolates were multidrug-resistant. It is estimated by predictive models that cephalosporin-resistant Enterobacteriaceae Escherichia coli and K. pneumoniae strains accounted for more than 5 million bloodstream infections and the carbapenem-resistant isolates were responsible for half a million cases, only in 2014 [12]. Certainly, the emergence of a more recent resistant clones has contributed to a much worse outcome.
Genomic Diversity, Antibiotic Resistance, and Prevention of K. Pneumoniae Infections
One of the challenges to control K. pneumoniae infections relies on the great genomic diversity of circulating strains. This species can colonize human, animal and even plant tissues, although little is known about key genes involved in host specialization. Moreover, K. pneumoniae displays the ability to exchange genetic material with other resident bacteria by using several traditional mechanisms besides less commonly regarded strategies, such as small plasmids [13]. Consequently, the panel of K. pneumoniae resistance is increasingly broader and include antibiotics of many families, as shown in Table 1 [14-16]. The construction of the table considered susceptibility test results provided by a piece of literature. Such tests aim to, following the isolation of a bacterial agent, determine the degree of sensitivity of the latter when in contact with one or more antimicrobials. From this test, the increase in virulence of a certain bacterium can be recorded by comparing the resistance of its already analysed strains. Quantitative susceptibility tests provide the value of MIC (Minimum Inhibitory Concentration). This value defines the minimum amount of antibiotic that is needed to inhibit the growth of the pathogen and serves as the basis for the resistance classification.
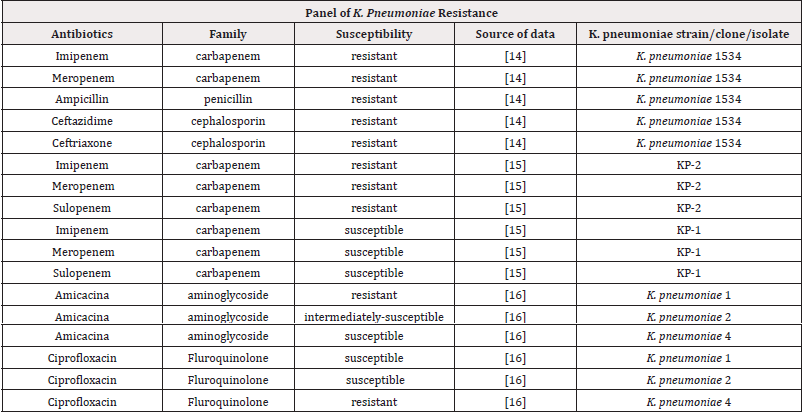
Table 1:Qualitative description of resistance of K. pneumoniae strains, considering different classes of antibiotics.
In the table, most of the results have already been presented qualitatively, indicating whether the K. pneumoniae strains analysed were resistant, intermediately susceptible, or susceptible. For quantitative results, which presented only the MIC value, the cut-off table provided by the BrCAST (Brazilian Committee on Antimicrobial Susceptibility Testing) was used. This committee is responsible for standardizing in vitro microbial susceptibility tests in Brazil. It is worth mentioning that the evaluation of the MIC value does not indicate the resistance mechanism. Despite this, the identification of the MIC has great value when related to clinical practice, since the knowledge of the minimum concentration to inhibit a microorganism could help to decide possible strategies for the therapy, preventing both its failure and the induction of new forms of resistance. The MIC value is also important as it is the only reliable method to phenotypically characterize certain bacteria and antibiotics, as qualitative methods may fail. In the case of K. pneumoniae, for antibiotics of the Phosphomycin, Tigecycline and Colistin classes, susceptibility can only be defined by quantitative processes, such as the MIC [17].
The data presented in the table were selected considering three factors: the diversity of antibiotics to which the bacteria were exposed, as shown for the K. pneumoniae 1534 strain; the difference in sensitivity among different strains found in the same patient, indicated by strains KP-1 and KP-2; and the difference in sensitivity between strains found in the same community, indicated by the strains K. pneumoniae 1, K. pneumoniae 2 and K. pneumoniae 4. Therefore, it is possible to analyse both the sensitivity of a strain, and the difference in sensitivity of strains of the same pathogen, which are coexisting in the same organism and/or in the same environment. This panel of antibiotic’s susceptibility unveils the genome-based K. pneumoniae versatility. In this sense, an open pangenome was found in a study that relied on the analysis of more than 300 isolate’s genomes. The authors affirm that additional genes will continue to be detected in future studies involving K. pneumoniae genomes [18], probably due to genome permissiveness besides the occurrence of horizontal gene transfer processes. These two factors-genome plasticity and ability to exchange DNA-could be the reasons, at least in part, of the increasing frequency of outbreaks of drug-resistant strains worldwide.
Particularly, the carbapenem-resistant infections highlight the limited pharmacological resources to manage the dissemination of bacteria as well as the clinical manifestations, besides the economic burden involved. Santos and Secoli (2019) [19] estimated a cost of more than US$ 4,000per patient infected with carbapenemresistant K. pneumoniae. Carbapenem-resistant strains have spread around the world, of which isolates derived from ST258 clone deserve close attention. Phylogenetic analysis demonstrated that the isolates descended from a single clone that diverged in capsular polysaccharide genes [20]. Based on the composition of these saccharides, K. pneumoniae strains can be classified in more than 70 serotypes. Capsular polysaccharide is a key virulence factor, involved in immune evasion by preventing the complement deposition. Other essential bacterial components in virulence and pathogenesis include: siderophores-molecules that chelate Fe-, lipopolysaccharide and pili-a motile element also known as fimbriae [21]. Given the emergence of multidrug-resistant K. pneumoniae strains and the high diversity of bacterial serotypes, vaccine-based prevention measures are urgently needed.
Vaccines are the main immunobiological tool to prevent pathogenic infections since the end of XVIII century, with Edward Jenner, Louis Pasteur, and Robert Koch studies, but just in the XX century, the technology needed to high scale production of the diverse formulations became widely available. Attenuated or inactivated microorganisms, protein subunits or toxoids, polysaccharides or glycoconjugates, coding DNA, ribosomal fractions and recently, recombinant adenovirus and mRNA formulations constitute diverse approaches to induce active and protective immune responses against human pathogens through vaccination. Many formulations have been proposed to prevent K. pneumoniae infections, including capsular polysaccharide-based strategies, an approach currently in use against diseases caused by other capsulated bacteria, such as Streptococcus pneumoniae. However, serotype diversity, based on capsular polysaccharide variation, T-cell independent response as well as the high cost represent a challenge to develop this kind of vaccine. For this reason, several cost-effective, T-cell dependent or high-coverage initiatives are currently being considered for clinical use, including capsular polysaccharide-protein carrier conjugates, protein-based formulations, and others [22].
Conclusion and Perspectives
Although much evidence has been accumulated, in the last years, about molecular mechanisms of pathogenesis, approaches to monitor emergent multidrug-resistant clones and to develop new strategies to prevent and control infections, especially caused by the invasive ones, this bacterium remains one of the least studied ESKAPE members. Furthermore, the paucity of global data regarding K. pneumoniae infections cases relies on the fact that this pathogen is not widely regarded as a compulsory notification issue yet. This scenario results in an underestimation of overall burden of this bacterium as a nosocomial- and community-associated superbug. In this sense, one might say that illnesses caused by this germ could be fairly classified as neglected diseases. Therefore, we can affirm that much investment and partnerships are still needed to address the burden of K. pneumoniae infections.
Acknowledgment
We would like to thank Federal University of ABC (UFABC) for institutional support and fellowship funding, in partnership with CNPq (Brazilian Council for Scientific and Technological Development), of Larissa A. Brigagão, PIC/PIBIC 01/2020 and 2021.
Conflict of Interest
We hereby declare that there is not a conflict of interest of any kind.
References
- Qiu Y, Lin D, Xu Y, Cheng Y, Wang F, et al. (2021) Invasive Klebsiella pneumoniae Infections in Community-Settings and Healthcare Settings. Infect Drug Resist 14: 2647-2656.
- Reid CB, Steele L, Pasquill K, Parfitt EC, Laupland KB (2019) Occurrence and determinants of Klebsiella species bloodstream infection in the western interior of British Columbia, Canada. BMC Infectious Diseases 19(1): 1070.
- Oliveira DMP, Forde BM, Kidd TJ, Patrick NA, Harris PNA, et al. (2020) Antimicrobial Resistance in ESKAPE Pathogens. Clin Microbiol Rev 33(3): e00181-19.
- Rice LB (2008) Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J Infect Dis 197(8): 1079-1081.
- Santajit S, Indrawattana N (2016) Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res Int pp.2475067.
- PAHO (2021) Americas report surge in drug-resistant infections due to misuse of antimicrobials during pandemic.
- WHO (2019a) Ten Threats to Global Health in 2019.
- CDC (2019) Antibiotic Resistance Threats in the United States pp: 150.
- Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR, (2019) Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front Microbiol 10: 539.
- WHO (2019b) The top 10 causes of death.
- Sands K, Carvalho MJ, Portal E, Thomson K, Dyer C (2021) Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nature Microbiology 6(4): 512-523.
- Temkin E, Fallach N, Almagor J, Gladstone BP, Tacconelli E, et al. (2018) Estimating the number of infections caused by antibiotic-resistant Escherichia coli and Klebsiella pneumoniae in 2014: a modelling study. Lancet Glob Health 6(9): 969-979.
- Ramirez MS, Iriarte A, Reyes Lamothe R, Sherratt DJ, Tolmasky ME (2019) Small Klebsiella pneumoniae Plasmids: Neglected Contributors to Antibiotic Resistance. Front Microbiol 10: 1-14.
- Yigit H, Queenan AM, Anderson GJ, Domenech Sanchez A, Biddle JW, et al. (2001) Novel Carbapenem-Hydrolyzing β-Lactamase, KPC-1, from a Carbapenem-Resistant Strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45(4): 1151-1161.
- Kaczmarek FM, Dib Hajj F, Shang W, Gootz TD (2006) High-Level Carbapenem Resistance in a Klebsiella pneumoniae Clinical Isolate Is Due to the Combination of blaACT-1 β-Lactamase Production, Porin OmpK35/36 Insertional Inactivation, and Down-Regulation of the Phosphate Transport Porin PhoE. Antimicrob Agents Chemother 50(10): 3396-3406.
- Ballot DE, Bandini R, Nana T, Bosman N, Thomas T, et al. (2019) A review of-multidrug-resistant Enterobacteriaceae in a neonatal unit in Johannesburg South Africa. BMC Pediatr 19: 320-329.
- Kowalska Krochmal, Dudek Wicher (2021) The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 10(2) 165: 1-21.
- Holt KE, Wertheim HH, Zadoks RN, Baker S, Whitehouse CA, et al. (2015) Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci 112(27): E3574-E3581.
- Santos WM, Secoli SR (2019) Economic burden of inpatients infected with Klebsiella pneumoniae Health Economics and Management 17(4): 1-8.
- DeLeo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, et al. (2014) Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci USA 111(13): 4988-4993.
- Ko KS (2017) The contribution of capsule polysaccharide genes to virulence of Klebsiella pneumoniae. Virulence 8(5): 485-486.
- Assoni L, Girardello R, Converso TR, Darrieux M (2021) Current Stage in the Development of Klebsiella pneumoniae Infect Dis Ther 10(4): 2157-2175.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.