Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Bacterial Extraction and Concentration from Blood through Filtration Processes
*Corresponding author: William G Pitt, Chemical Engineering Department, 330 Engineering Building, Brigham Young University, Provo, Utah, USA 84602, Tel: 801-422-2589; Fax: 801-422-0151; Email: pitt@byu.edu
Received: August 01, 2023; Published: August 28, 2023
DOI: 10.34297/AJBSR.2023.19.002659
Abstract
Blood stream infections (BSIs) are challenging to rapidly and precisely diagnose. Current clinical diagnostic methods for BSI identification require culturing the blood sample for many hours prior to identifying the bacteria and possible antibiotic resistance. Removing the culturing step from the bacterial identification process of a BSI provides a significant reduction in the processing time, but shifts the rate-limiting step from the growth time to the concentration procedure, since CFU levels in clinical blood samples can be as low as 10 CFU/mL in blood. This study developed a novel whole blood filtration method to concentrate bacteria from a BSI without culturing the blood or bacteria. Filtering whole blood achieved 100% bacterial removal from 5mL of whole blood in ~90 s, but the bacteria were difficult to remove from the filter. Bacterial removal from the filter after blood filtration was also investigated. At high laboratory loadings of bacteria into freshly collected human blood, a blood lysis solution of 3% Tween 80 followed by a 3% Pluronic F108 backflush solution achieved 80% removal of the bacteria from the filter. This is a major step in collection of bacteria directly from blood for rapid diagnostics without a culture amplification step and with a single backflush.
Keywords: Blood; Bacteria; Separation; Filtration; Rapid Diagnostics
Abbreviations: BSI: Blood Stream Infections; RBC: Red Blood Cell; WBC: White Blood Cell; SDS: Sodium Dodecyl Sulfate; Gu+: Guanidine; Hct: hematocrit; CHAPS: 3-[(3-cholamidopropyl); dimethylammonio]-1-propanesulfonate; PCTE: Polycarbonate Track- Etched; IRB: Institutional Review Board; PVP: Polyvinyl Pyrrolidone; EDTA: Ethylenediaminetetraacetic acid
Highlights
I. Bacteria recovered from lysed whole blood by filtration
II. Various chemical formulations for filtration of 5 mL blood
III. Recovered bacteria are backflushed from filter
IV. Recovered bacteria are viable for further processing
V. Filtering and recovery take about 3 minutes
Introduction
The rise of antibiotic resistant bacteria threatens worldwide health and elevates the need for rapid identification of species and antibiotic resistance profiles of pathogens, especially in bloodstream infections (BSI) [1]. BSIs can produce clinical sepsis in patients, even at very low bacterial concentrations (1 to 100 CFU/mL) [2,3]. The survival rate of some BSI-induced sepsis infections can decrease by as much as 7.6% per hour of ineffective treatment [4,5]. Unfortunately, current clinical diagnostics require growth amplification for identification [6], resulting in diagnostic procedures that require at least 12-24 hours (usually longer) between sample acquisition and reporting [6,7]. These studies have shown that timely administration of efficacious antibiotic regimens decrease the mortality rates for BSIs [8,9]. Thus, reducing the diagnostic time (sample acquisition to reporting) can greatly reduce mortality and morbidity.
The need to decrease the BSI diagnostic time has promoted development of novel technologies and devices to directly collect bacteria from blood [10]. Our proposed process described herein improves upon direct blood-lysis filtration technologies [11-14]. The basis of this method is the lysis of formed blood elements (red blood cells (RBC), white blood cells (WBC), and platelets) through the use of detergents, salts, and/or proteases followed by the filtration of the solution for isolation and concentration of bacteria.
Since blood-lysis filtration is not new, a review of past research shows how the present study differs. For comparison of different lysing procedures and lysis solutions (between published and our experiments), we devised a ratio that compares the volume of whole blood that was actually filtered to the filterable area of the filter. This ratio does not include any dilution of the blood by the lysis solution; for example using a filterable area of 1 cm2, if 5 mL of whole blood were mixed with 15 mL of a lysis solution, the ratio would be 5 mL/cm2 of whole blood per filter area, as long as the entire 20 mL of the blend was filtered. However, if only 10 of 20 mL blood-lysis blend were passed before clogging the filter, then the ratio would be 2.5 mL/cm2 of whole blood per filter area.
Initial publications regarding this technique described whole blood filtration after dilution with and lysis by a solution of Triton X-100 and Na2CO3 [13,14]. The best results were 0.288 mL/cm2 of blood per filter area. When proteases were incorporated into the solution to further improve the process, the ratio improved to 0.865 mL/cm2 [14,15]. However, this lysis solution was toxic to some bacteria if not filtered within minutes. Less toxicity was observed with Tween 20 [16]. Blood and detergent concentration, incubation time, and solution pH play important roles in blood cell lysis [17]. 100% lysis was achieved using 0.7% Tween and 1/10 diluted blood incubated for 15 min at pH >10, or 30-min incubation at pH=9, or 90-min incubation at pH 6-8. The use of proteases made further improvements [17]. Obviously, this is a complex and multi-faceted problem of protein and lipid aggregates blocking filter pores.
There are other studies that investigated altering the filter membrane surface to ensure bacterial collection, [18] but more importantly for rapid identification is subsequent removal of bacteria from the filter. Zierdt examined bacterial removal from filters and found only 12% of the bacteria were removed by backflushing using 5 mL of phosphate buffer [14]. Detergent was needed to remove bacteria and only 45% of bacteria could be removed with a single backflush [19]. Pre-soaking the filter in the detergent provided a 2.5- to 5-fold increase in bacterial removal. 100% removal was obtained using multiple elution volumes or recirculation.
Furthermore, salt effects on proteins reveal that salt chemistry and concentration are key parameters. Salts can be either chaotropic or kosmotropic. Chaotropes, such as guanidinium chloride, interfere with hydrophobic interactions and thus destabilize proteins [20]. Kosmotropes, such as magnesium sulfate, stabilize proteins and hydrophobic aggregates. NaCl is a weak kosmotrope and has a stabilizing effect (i.e., prevent precipitation) on detergent-protein and detergent-protein-lipid complexes [21]. Furthermore, salt concentration was important in stabilizing aggregates in sodium dodecyl sulfate (SDS), with increasing salt concentrations leading to larger micelle aggregates for both NaCl and CaCl2 [22]. For salts of different anions and cations, the preferential protein hydration generally follows the Hofmeister series: CO3 2- > SO4 2- > COO- > HCO3- > Cl- > I- > NO3- > ClO4- > SCN- and Zn2+ > Mg2+ > Cu2+ > Ca2+, Ba2+ > Li+ > Na+ > K+ > Mn2+, Ni2+ > NH4+ > Gu+ (Gu+ = guanidine) [23,24]. These relationships are also dependent on salt concentration and solution pH [25]. Preferential hydration means the protein prefers to be surrounded by water, usually maintaining protein function (stability), while preferential binding means that the protein prefers to be surrounded by the salt which causes destabilization and conformational changes to the protein [26]. At a certain salt concentration, preferentially hydrated proteins maintain a liquid layer that is no longer in equilibrium with the bulk solution causing the protein to precipitate; preferentially bound proteins can transition from preferentially bound to preferentially hydrated as the protein unravels, which also may cause the protein to precipitate [26,27].
As for blood, protein-containing cell membranes in the presence of non-ionic detergents were never completely solubilized and contained large particles between 0.5-1 μm in diameter [28]. However, this was not the case when using SDS as it completely solubilized both proteins and lipids [28]. Factors which affect membrane solubilization include detergent chemistry, pH, ionic strength, temperature, and the size of the micelles formed by the detergent [29]. High molecular weight micelles form larger structures, which when complexed to proteins or membrane fragments, have more potential to clog filters.
In studies regarding Triton X-100 interactions with blood at very low hematocrits of 0.15, 0.30, and 0.45 (vol %), 213-μM and 255-μM Triton produced 100% hemolysis for 0.30 and 0.45 Hct levels, respectively [30]. Comparable results were reported for non-ionic detergents, Brij 58 and Brij 98, against Hct levels of 0.15, 0.30, 0.45 and 0.60 vol%. Blood with hematocrits of 0.30, 0.45 and 0.60 could be completely lysed using Brij 58 at concentrations of 37.6, 43.1 and 47.0 μM, respectively [31]. Furthermore, lower concentrations of Brij 98, 20.4, 22.2 and 25.1 μM, were needed for complete hemolysis of blood with 0.30, 0.45 and 0.60 Hct levels, respectively. Another study examined polyoxyethylene alkyl ethers, from C10E8 to C18E8 and found that complete hemolysis could be achieved at Hct = 0.45 [32].
In addition to direct whole blood lysis, our present study also examined doing blood-lysis filtration to collect bacteria from plasma from whole blood spun upon a previously described centrifugal- sedimentation device [10,33-37]. A lysis solution is often needed for the plasma recovered after spinning on this device because the device does not separate all RBCs from the resulting plasma.
In general, large blood dilution volumes lead to large waste volumes, which clinics would rather avoid. Therefore, our study sought a filtration procedure for a 1:1 blood: lysis solution that would work for both sedimentation-recovered plasma and for whole blood. Unfortunately, a 1:1 blood: lysis solution for whole blood has not yet been found; but a working 1:9 blood: lysis solution for whole blood was achieved. Because this bacterial extraction method is designed to be combined with a clinical rapid identification assay, small final volumes were desirable, so 25-mm-diameter polycarbonate track-etched (PCTE) filters were used. A second goal of this study was to filter bacteria from 5 mL of whole blood; our best formulation yields a whole blood per filterable area ratio of 1.43 mL/ cm2, greatly exceeding previous studies.
Methods
Two main sets of experiments were performed in this study. The first set involved investigating the effects of different lysis solutions on whole blood suspensions and plasma suspensions (recovered after spinning) without bacteria present. These experiments analyzed different blood-to-filter-area ratios with the goal of obtaining the highest ratio. The second set of experiments involved investigating the effects of different lysis and backflush solutions on the recovery of bacteria from the filter surface. These experiments filtered a standardized blend of a blood-bacteria suspension and a lysis solution as described below.
Filtration only
Both whole blood and plasma recovered from “disk spinning” were investigated for their ability to be filtered after lysing the blood cells present. During the evaluation of different lysis solutions, a 25-mm track-etched filter with 0.8-μm pore diameter (#PCT0825100, Sterlitech) was used. Before filtering the blood, a lysis solution was made consisting of various salts and detergents in distilled water. Various amounts of blood (whole or recovered plasma) and lysis solution were blended to evaluate the filtration of different blood volumes at different dilution ratios. The blend of the blood suspension and lysis solution was pushed via syringe (3 mL, 5 mL, or 10 mL depending on blend volume) through the filter using a reusable syringe filter holder (#EW-06623-32, Cole Palmer). The volume of blend that could be filtered by hand, called filtrate, was recorded and the blood to filterable area ratio calculated. Human blood was obtained under an IRB-approved protocol at Brigham Young University (IRB #X18340) from volunteers by venipuncture and collection into EDTA-containing vacutainers (BD#366643, Becton Dickinson), and was used the same day.
Bactericidal testing

Figure 1: Bactericidal effect of SDS upon E. coli at 10 min and 1 hr of incubation. Error bars represent 95% confidence intervals.
Toxicity due to SDS (#L4509, Sigma Aldrich) was measured by dissolving 4.5 g into distilled water and then adding distilled water to the vial once all the SDS was dissolved, to a total of 50 mL. Then serial dilutions were made from this stock solution to formulate the desired test concentrations. Bacteria used for all experiments were Escherichia coli (strain DH5α), streaked from frozen stock and grown overnight with shaking in nutrient broth. Nine mLs of bacteria were removed from the broth in 1 mL aliquots which were then spun down in a Horizon 642E centrifuge (#22-029-375, Fischer Scientific) at 3328 rpm for 10 min, and the supernatant removed. Then 1 mL of the desired SDS concentration was pipetted into each vial and the bacteria resuspended. The viable bacterial concentration was determined by colony plate counting after exposure for 10 min or 1 hr to reveal the bactericidal effects (see Figure 1). Plate counting was done by removing 100 μL and performing five 10-fold serial dilutions; then 50 μL of the 10-2 to 10-5 dilutions were plated and the plates incubated for 24-48 hours before counting colonies.
Filtration and backflush
For experiments involving backflushing the bacteria from the filters, there are 3 fluids involved: a bacterial deposition fluid; a lysis solution; and a backflush solution. The types of bacterial deposition fluids are 1) bacteria in PBS (control without blood components); 2) bacteria in plasma from spun blood (~60% of original platelets and ~0.5% of original RBCs); 3) bacteria in 1% RBCs suspended in PBS; and 4) bacteria in whole blood. To make 1% RBCs, whole blood was centrifuged in centrifuge tubes in a Horizon 642E centrifuge (#22-029-375, Fischer Scientific) at 3328 rpm for 10 min to spin down the blood cells. The plasma and buffy coat were removed, and the remaining RBCs were washed in PBS and recentrifuged. One milliliter of the RBC pellet was then added to 99 mL of PBS to make the 1% RBC suspension. Bacteria used for all experiments were Escherichia coli (strain DH5α), streaked from frozen stock and grown overnight with shaking in nutrient broth. In these experiments, bacteria were washed in PBS before dilution and inoculation into the desired bacterial suspension fluid at a concentration of 2 x 107 CFU/mL.
The formulation of the lysis solutions was standardized as follows. The salts and detergents for the lysis solution were dissolved in distilled water to a final volume of 45 mL. The solution was vortexed and heated to 50℃ to ensure dissolution. After cooling to room temperature, 5 mL of the bacteria suspension – in either PBS, spun plasma, 1% RBCs suspended in PBS, or whole blood – was added to the filtering solution and mixed by vortexing. The blend was then filtered until either the filter was clogged or the entire 50 mL of blend was filtered. The volume of blend that passed through the filter, called filtrate, was recorded. If the entire blend volume was filtered, 3 mL of PBS were additionally pushed through to wash the filter before backflushing.
To evaluate the different backflush solutions, two 3-mL syringes were used, and the backflush solution was “reverse filtered”, or pushed through the filter in the opposite direction of the flow of the blend. The first syringe, labelled “A”, pushed 2 mL of the backflush solution through the filter into the second syringe, labelled “B”. Syringe “B” had been weighed empty and was weighed again after collection of the backflush filtrate to determine the backflush volume and eventual bacterial concentration. The filters were a 25- mm track-etched filter with 0.4-μm pore diameter with either a hydrophilic (PVP-coated, #PCT0425100, Sterlitech) or hydrophobic (uncoated, #PCTF0425100, Sterlitech) surface. The pore size of these filters was smaller than for filters used during the development of the filtration solution because it was observed that during filtration experiments that 0.8-μm filters allowed about 5% of the E. coli to pass through the filter.
To quantitate the bacterial recovery from backflushing, the total bacteria delivered to the filter and the total bacteria removed from the filter were calculated. The ratio of the total bacteria removed to the total bacteria delivered provides the recovery fraction of the bacteria. This value multiplied by 100% gives the percent recovery for the experiment. Total bacteria delivered was calculated by multiplying the bacterial concentration by the volume of the bacteria-blood suspension that was filtered. The volume of the bacteria-blood suspension was determined by sucking up into a syringe approximately 5 mL of the bacteria-blood suspension and weighing it, emptying the syringe into the lysis solution, weighing the empty syringe, and then multiplying the difference in the weights by the 1.061 g/mL (the approximate density of whole blood). The bacterial concentration of the bacteria-blood suspension was determined by plate counts. The percentage of the blend that was filtered was calculated by dividing the amount of filtrate by 50 mL (the total volume of the blend).
The total bacteria removed from the filter was calculated by multiplying the bacterial concentration by the volume of the backflush filtrate in syringe “B”. The volume of the backflush filtrate in syringe “B” was determined by multiplying the difference between the weights of syringe “B” empty and filled with the backflush filtrate by the density of the solution (densities ranged from 1.004 g/mL to 1.009 g/mL). The bacterial concentration of the backflush filtrate was determined by plate counting.
Statistics
The statistical significance of experiments was analyzed using Welch’s t-test (unequal variances and sample sizes) with statistical significance being p < 0.05.
Results
Filterability of lysed blood suspensions
The main difference between filtering whole blood and filtering plasma recovered from spinning is the number of blood cells present [10,34,35]. Plasma recovered from spinning usually has < 1% of the original RBC concentration and an even lower percentage of WBCs [34], resulting in a significantly smaller formed element membrane area to be lysed. Thus, when evaluating lysing solutions, emphasis was placed upon a solution’s ability to accomplish the more difficult task of lysing and filtering a whole blood suspension.
SDS is an anionic surfactant commonly used to disrupt cell membranes and proteins. It is also known to be bactericidal. Therefore, the bacterial survival in varying SDS concentrations was first analyzed. Figure 1 shows that concentrations at or above 0.111% (w/v) SDS were bactericidal to the E. coli under these evaluation conditions. Based upon these results and the postulate that there may be other bacterial species more sensitive to lysis by detergents than is E. coli, different SDS concentrations were evaluated to produce filterable suspensions of whole blood and suspensions of plasma recovered from spinning. The opaqueness of the whole blood when mixed with 0.111% SDS lysis solution (image not shown) suggested that not all blood cells were lysed; thus, higher concentrations of SDS were evaluated to see if complete lysis and filtration were even possible in whole blood using only SDS. Table 1 shows the results of the SDS lysis solutions both for plasma recovered from spinning and for whole blood. Significant filtration was achieved only when the SDS concentration exceeded 6.7%. If viable bacteria are needed, this concentration cannot be tolerated.

Table 1: Lysis and filtration of plasma and whole blood at various SDS concentrations.
* This percentage of the suspension was successfully filtered.
‡ Because the plasma suspension has significantly reduced blood cell counts, the ratio is not reported for plasma suspensions.

Table 2: Effects of a protease upon lysis and filtration of plasma and whole blood in a saponin solution.
* This percentage of the suspension was successfully filtered.
‡ Because the plasma suspension has significantly reduced blood cell counts, the ratio is not reported for plasma suspensions.
† Protease from Aspergillus melleus.
Another detergent used to disrupt cell membranes is saponin. Unlike SDS, saponin is a non-ionic surfactant and a less potent bactericidal compound. Zierdt tested saponin and found it to be comparable to Tween 20 as a blood cell lysis compound [16]. However, Zierdt also used a protease when evaluating his blood lysis solutions. Table 2 shows our similar data of lysis and filtration of blood cell-lysis blends with and without proteinase from Aspergillus melleus, both for plasma recovered from spinning and for whole blood. Table 2 reveals the importance of having a protease present in the lysing solution. For example, comparison of experiments 1 and 4 of Table 2 show that adding 0.33 mg protease to the 45 mL of lysis solution increased filterability from 4% to 100%. 100% filtration is marked in red for emphasis.
However, proteases can be expensive, and many require specific temperatures or incubation times to be effective. Therefore, our subsequent research followed the publications of Farmer et al. [13] and Sullivan et al. [38], since they achieved whole blood lysis and filtration without the use of a protease. In our studies without proteases, the focus was on achieving lysis with a small blood to lysis solution ratio, and various salts were explored for their effect upon blood lysis and filtration. Table 3 presents the salts (1M concentrations) used in combination with Triton X-100 and how well they lysed and promoted filtration of 5 mL of whole blood mixed with 45 mL lysis solution (1:9 blood:lysis ratio).

Table 3: Whole blood filtration percentages using Triton X-100 and different salts at 1:9 blood dilution.
* This percentage of the blood-lysis-solution blend was successfully filtered. Red color indicates the blends that produced 100% filtration, which is a blood-to-filterable-area ratio of 1.43 mL/cm2.
The filtration results indicate that there is an interplay between the concentration of the detergent and the concentration of the salt that is dependent on the chemical character of the salt. In general, lysis solutions formulated with 4% Triton X-100 promoted better filtration than solutions with 2% or 6% surfactant. Whenever a lysis solution of 4% Triton X-100 and 1M salt produced 100% filtration, that particular salt was further examined at 0.5 and 2.0 M concentration with 4% Triton X-100. Table 4 shows the results of experiments of those particular salts at different salt concentrations and 4% Triton X-100. These results confirm that the ability of a detergent-salt solution to promote filtration of whole blood is dependent on the detergent concentration, the salt concentration, and the chemical makeup of the salt. In general, filterability using hydroxide and carbonate salts was sensitive to the salt concentration, while filterability with sodium and potassium chloride was insensitive to this range of salt concentrations (0.5M to 2M).

Table 4: Whole blood filtration using 4% Triton X-100 with different salt concentrations at 1:9 blood dilution.
* This percentage of the blood-lysis-solution blend was successfully filtered. Red color indicates the blends that produced 100% filtration, which is a blood-to-filterable-area ratio of 1.43 mL/cm2.
The results from Table 4 for sodium carbonate suggest that there is some variation in filterability of a whole blood blend at different blood dilutions. Prior publications of whole blood lysed with a 0.225:9 ratio of blood to 3.8 mM Na2CO3 in 0.05% Triton-X-100 [13] and a 0.393:9 ratio of blood to 38 mM Na2CO3 in 0.025% Triton- X-100 [38] produced solutions that were completely filterable. However, in our hands, these mixtures did not produce completely filterable solutions. A possible contributing difference is that blood in our lab was collected into EDTA anticoagulant, and in these prior publications blood was collected into polyanethol sulfonate. Also, our solution only had a 1minute incubation time while the incubation time of the prior solutions with blood lasted until clearing was apparent.
Our next set experiments further explored the effect of salt concentration. Sodium bicarbonate, sodium chloride and sodium acetate salts were used to compare results from a basic, neutral, and acidic salt, respectively. Table 5 shows the results of these experiments with differing Triton X-100 concentrations and with various concentrations of each of the 3 salts at a 1:9 whole blood blend for 5 mL of whole blood. These data show that the filterability using the neutral salt (NaCl) is less sensitive to salt and surfactant concentration than either the acidic or basic salts. Not all of the different salt and Triton X-100 solutions that were combined with whole blood at a 1:9 ratio were successful in producing 100% filterability of the blend. Of those combinations that produced 100% filtration, there are 9 combinations using sodium chloride, while there are only 3 sodium bicarbonate combinations and 2 sodium acetate combinations.
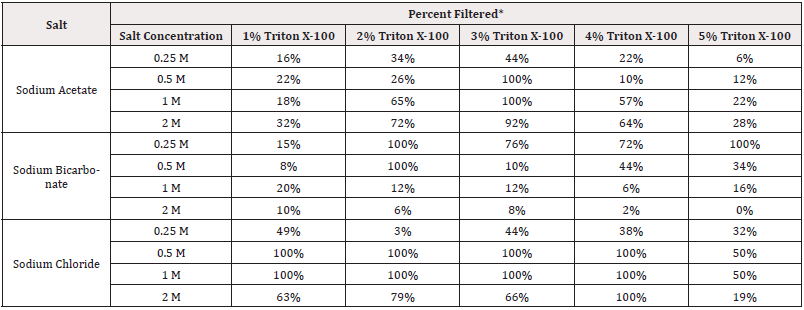
Table 5: Results of whole blood filtering varying Triton X-100 and salt concentrations for an acidic, basic, and neutral salt.
* This percentage of the blood-lysis-solution blend was successfully filtered. Red color indicates the blends that produced 100% filtration, which is a blood-to-filterable-area ratio of 1.43 mL/cm2.
All of the previous blood-lysis-blend filtration experiments were performed using a blood Hct between 42 and 46, which is within the normal physiological range. However, this technology will be used in diagnosing sick people, whose hematocrit levels could be abnormally high due to dehydration and other factors [39]. Therefore, filtering whole blood with exceptionally high Hct was also examined by adding RBCs to the donated blood to raise the Hct to 65. While this is an unrealistic Hct, even for an ill person, it was postulated that a solution that would allow 65% hematocrit blood to be filtered would be useful for all Hct levels encountered in a clinical setting. The 14 salt and Triton X-100 combinations that allowed 100% of the whole blood blend to be filtered (see Table 5) and two other combinations (0.5M and 1 M Sodium Chloride with 5% Triton X-100) were studied to see if they promoted successful filtration of a 1:9 dilution of 65% hematocrit whole blood blend. Similar to all previous whole blood filtration experiments, 5 mL of the 65% hematocrit whole blood was filtered through a 0.8-μm filter. Table 6 shows the results for the filtration of high Hct whole blood blend. Only 3 of the salt and Triton X-100 combinations with 65% hematocrit blood were filterable: 0.5 M sodium bicarbonate with 2% Triton X-100, 0.5 M sodium chloride with 4% Triton X-100, and 1 M sodium chloride with 5% Triton X-100.

Table 6: Results of testing combinations of Triton X-100 and sodium carbonate, sodium chloride, or sodium acetate on 65% hematocrit blood.
* This percentage of the blood-lysis-solution blend was successfully filtered. Red color indicates the blends that produced 100% filtration, which is a blood-to-filterable-area ratio of 1.43 mL/cm2.

Table 7: Results of whole blood filtering using a 4% Triton X-100 and 0.5 M NaCl lysing solution at hematocrit levels of 55%, 58%, 65%.
* This percentage of the blood-lysis-solution blend was successfully filtered. Red color indicates the blends that produced 100% filtration, which is a blood-to-filterable-area ratio of 1.43 mL/cm2.
Because of the low number of salt and Triton X-100 combinations that produced completely filterable blends when combined with blood at Hct = 65, the influence of Hct on filterability was studied by using the salt and Triton X-100 combinations at 3 different Hct levels in further filtration experiments. These Hct levels were 55, 58 and 65. For these experiments, sodium chloride was chosen as the salt. In addition to varying the Hct of the blood, the sodium chloride concentration and Triton X-100 concentration were varied slightly to determine the sensitivity of the filtration of the sample. Based on our previous experiments, it was hypothesized that both Hct and small variations in both the salt and detergent would affect the filterability of the blood. The results of these experiments (see Table 7) show that the 0.5 M sodium chloride and 4% Triton X-100 combination is very robust with respect to variable Hct, as it produced completely filterable solutions when combined with blood having Hct of ~45 (see Table 5), 55, 58, and 65. Furthermore, the results also show the sensitivity in filtration to small changes in salt concentration. However, this sensitivity is not seen with deviations in detergent concentration.
In general for these and previous combinations in which 5 mL of blood and 45 mL of lysis solution were successfully filtered, the mixing and filtering process took only about 2 to 3 minutes. The mixing and vortexing was 10 to 20 seconds, followed by loading syringes and filtration of 2 minutes, or slightly more. With preparation and practice the filtration could be done in about 150 seconds.
Backflushing
In designing experiments to measure and optimize the removal of bacteria from filters by backflushing, we hypothesized that the presence of multiple layers of bacteria stacked on the filter would yield higher removals since only the bottom layer would interact with the membrane surface. Therefore, experiments were designed to identify the optimum backflush solution when there was a single layer or less of bacteria on the filter. The filters used were 25-mm-diameter track-etched filters with 0.4-μm pores (for details see Filtration and Backflush method section). According to the manufacturer of the syringe filter holder, the filtration area of the holder is 3.5 cm2. E. coli bacteria were modelled as approximately 3.5 μm long by 1 μm wide for an area of 3.5 μm2. This means that a single layer of side-by-side bacteria on the filter would contain approximately 1 x 108 CFU. Therefore, to ensure less than single layer coverage, these experiments employed not more than 108 CFU per membrane.
Two sets of control experiments were performed with the bacteria suspended in PBS. The first control experiment (only bacteria) consisted of filtering only 2 mL of a bacterial suspension of 2 x 107 CFU/mL, without blending the bacterial deposition fluid with the lysis solution. The second control experiment (bacteria in lysis solution) used 5 mL of the bacterial suspension in PBS and blended it with the best filtration lysis solution from the filtering experiments: 4% Triton X-100 with 0.5 M NaCl. The backflush solutions (n=6 for all solutions) examined were pure water, a 3 M NaCl solution (high salt concentration), a 0.9% (w/v) NaCl solution (low salt concentration, physiological saline), a 3% (v/v) Triton X-100 solution (high detergent concentration), and a 0.5% Triton X-100 solution (low detergent concentration). Triton X-100 was examined since it was the detergent used in the lysis solution developed through the previous filtration experiments. It was hypothesized that either of the detergent solutions would adequately remove the most bacteria with the best removal achieved by the higher detergent concentration. However, the results revealed that the hypothesis was incorrect with pure water achieving the best backflush solution for the bacteria-only control and the low salt solution achieving the best backflush solution for the bacteria-lysis-blend control.
For the bacteria-deposited-from-water control, backflushing with pure water achieved an average bacterial removal of 59.7%, while the low and high detergent solutions only achieved average bacterial removals of 3.9% and 4.9%, respectively (see Figure 2A). The salt solutions were comparable to each other with the low salt solution achieving an average bacterial removal of 48.7% and the high salt solution achieving an average bacterial removal of 49.5%.
For the blend of bacteria with the Triton-X-100 and NaCl lysis solution, the low salt solution achieved an average bacterial removal of 32.7%, while pure water only achieved an average bacterial removal of 0.9% (see Figure 2B). The high salt solution and the low detergent solution achieved similar average bacterial removals of 8.2% and 8.1%, respectively, while the high detergent solution only achieved an average bacterial removal of 1.4%.
The surprising backflush removal of bacteria deposited from water (Figure 2A) seems to be in contrast to the results found by Zierdt [19]. However, the backflush removal of bacteria deposited from the mixture of Triton-X-100 and NaCl are consistent with the results found by Zierdt. This observation prompted us to further investigate the role of the deposition solution suspending the bacteria as well as the backflush solution. Three additional detergents (Tween-20, Pluronic F108, and Brij 58) were added to the set of backflush solutions employed for the bacteria-lysis blend, at both a low concentration of 0.5% and a high concentration of 3%. More backflush removal was observed using these surfactants–on the order of 50% for the Tween and Pluronic backflush solutions and about 80% removal for 3% Brij 58 (see Figure 2B).
Additional experiments were performed by depositing bacteria from a 10% lysed blood solution (5 mL of whole blood suspended in 45 mL of 4% Triton X-100 with 0.5 M NaCl) to determine whether the presence of any detergent-salt chemistry or the lysed blood cell components were inhibiting the removal of bacteria from the filter. Figure 2C shows these results (n=6 for each combination of suspension fluid and lysis fluid) which reveal that the lysed blood solution is associated with high bacterial retention on the filter. When depositing bacteria from the 10% lysed whole blood solution, the backflush solutions of pure water, low and high salt, and low and high Triton X-100 produced minimal bacterial removal (no more than 2%). The low Tween-20, Pluronic F108, and Brij 58 solutions achieved average bacterial removals of 3.0%, 6.6% and 23.1%, respectively; similarly the high Tween-20, Pluronic F108, and Brij 58 solutions achieved average bacterial removals of 2.3%, 16.4% and 16.5%, respectively. Figure 2D shows comparisons of bacterial removal after deposition from the 3 types of deposition suspensions.

Figure 2: Percent bacterial removal by backflushing when applying the bacteria to the filter in different mediums: A) bacteria deposited from pure water and removed by various backflush solutions; B) bacteria deposited from 4% Triton X-100 with 0.5 M NaCl and removed by various backflush solutions; C) bacteria deposited from 10% lysed blood and removed by various backflush solutions; D) comparative graph of the results from similar backflush solutions used in A-C. The backflushing solutions are listed on the y-axis. The x-axis is the percentage of E. coli removed from the filter. Error bars represent 95% confidence intervals (n ≥6).
Since the presence of lysed blood cell components appeared to be the greatest factor inhibiting the removal of bacteria from the filter, the components of lysed blood cell were further investigated by comparing to experiments using either plasma (with bacteria) recovered from spinning or a 1% RBCs in PBS suspension. The plasma recovered from spinning had a small Hct of about 0.5 and maintained approximately 60% of the platelets and almost all the plasma proteins. The 1% RBCs suspension only contained RBCs and little if any protein. These were blended 1:9 into the best lysis formulation of 4% Triton-X-100 and 0.5 M NaCl. Because of the difference in RBC counts and the difference in the presence of platelets and plasma proteins between the two suspensions, the impact of the different blood components on the bacterial removal could be evaluated. Bacterial deposition from each of these suspensions were challenged by backflush solutions with high and low concentrations of Triton X-100, Tween-20, Pluronic F108, and Brij 58; see Figure 3.
The data shown in Figure 3 reveals that deposition from the plasma suspension and the 1% RBC suspension produced similar retention on the filter. However, even acknowledging the scatter in the data, in general the 1% RBC suspension produced more retention on the filter overall than did the plasma suspension. Therefore for further optimization of bacterial removal from the filter, subsequent experiments were focused on challenging the most adhesive deposition solution: a bacteria-blood-lysis blend made from the 1% RBCs. In addition to testing backflushing solutions, subsequent experiments included evaluating different detergents in the deposition solution and evaluating different types of filters in search of the optimum combination. The different detergents evaluated for the lysis solutions were 1) a combination of 1% Brij 58, 1% Tween-80, and 1% Pluronic F108 and a similar combination at half those concentrations; 2) Pluronic F108 at 3% and 1% (w/v); 3) Tween-80 at 3% and 1% (v/v); 4) Tween-20 at 3% and 1% (v/v); 5) Brij 58 at 3% and 1% (w/v); and 6) Triton X-100 at 3% and 1% (v/v). Additionally, these blood lysis solutions also contained 0.5 M NaCl.
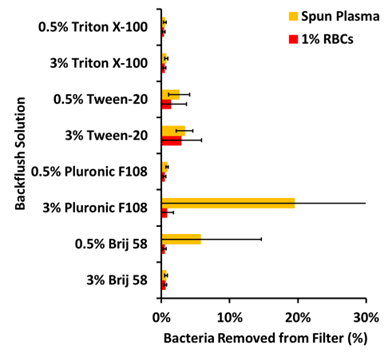
Figure 3: Percent bacterial removal by backflushing when using either 1% RBCs in PBS or recovered plasma after spinning mixed with lysing solution as the medium for bacterial filtering. Error bars represent 95% confidence intervals (n=6).

Figure 4: Percent bacterial removal (mean values, n=4) from hydrophilic filters (PVP-coated) when employing added detergents in the blood lysis and deposition solution with 1% lysed RBCs (in PBS) and then challenging with different backflushing solutions. Solid colours (panel A) represent non-pre-soaked filters and dashed colours (panel B) correspond to filters pre-soaked filters in same backflush solution. The entries on the y-axis indicated the various deposition (lysis) solutions. The colours indicated the types of backflush solution.
In this set of experiments, two different types of filters were employed: 1) 0.4 μm hydrophilic track-etched filter (polycarbonate filter with a polyvinylpyrrolidone (PVP) surface-coating) and 2) 0.4 μm hydrophobic track-etched filter (polycarbonate filter with no surface-coating). The backflush solutions were 3% Brij 58, 3% Tween-20, 3% Tween-80, and 3% Pluronic F108 (n=4 for each lysis solution and backflush solution combination). Figure 4 shows the removal efficiencies for each detergent combination using the hydrophilic filter, while Figure 5 shows the removal efficiencies for each detergent combination using the hydrophobic filter.
There are some significant observations relating to this set of experiments. First, there is a general trend that more bacteria are removed from hydrophobic filters, and slightly better removal occurs when the filter is presoaked in the backflush solution. Secondly, there are some lysis deposition solutions that do not allow significant removal, such as Triton-X-100 on hydrophilic filters, and Tween-20 on hydrophobic filters. On the other hand, there are some lysis deposition solutions that in general allow more recovery than others, such as the 3% Brij 58 on pre-soaked hydrophobic filters. As for backflush solutions, the 3% Brij 58 tends to accomplish more bacterial removal than the 3% Tween-20, 3% Tween-80, or the 3% Pluronic F108.
In generally the backflushing can be done fairly quickly, in about 30 seconds or less.
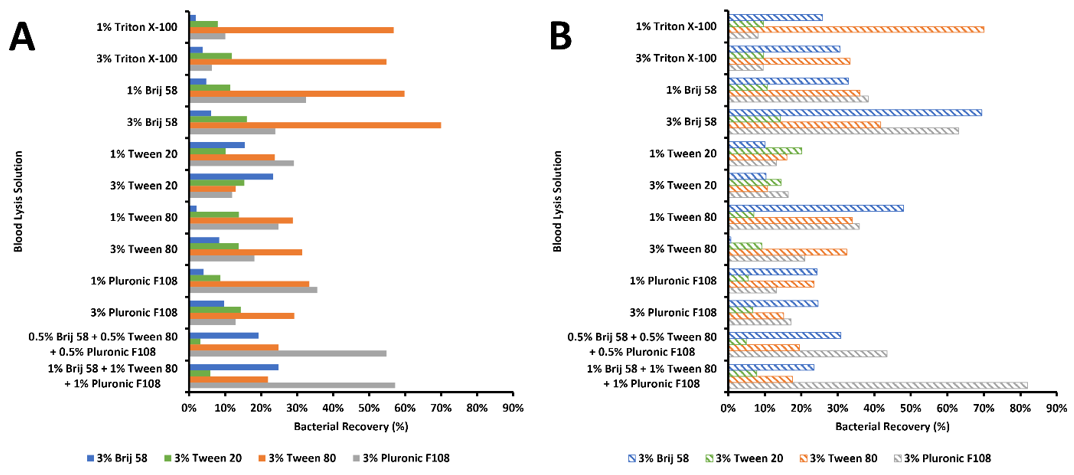
Figure 5: Percent bacterial removal (mean values, n=4) from hydrophobic filters (PVP-free) when employing added detergents in the blood lysis and deposition solution with 1% lysed RBCs (in PBS) and then challenging with different backflushing solutions. Solid colours (panel A) represent non-pre-soaked filters and dashed colours (panel B) correspond to filters pre-soaked filters in same backflush solution. The entries on the y-axis indicated the various deposition (lysis) solutions. The colours indicated the types of backflush solution.
Discussion
Removing bacteria from blood and plasma
Using filters to recover bacteria directly from blood or plasma is a challenging task. Removing bacteria from blood by size-exclusion filtration requires the blood cell membranes to be fragmented such that they are small enough to pass through the filter pores. According to Jones [41] there are four steps in the interaction between detergents and cellular membranes: 1) detergent accumulates in the membrane until saturation, 2) additional detergent lyses the membrane, 3) additional detergent solubilizes the membrane, releasing lipid-detergent micelles and protein-lipid-detergent complexes, and 4) additional detergent then releases protein-detergent complexes from the protein-lipid-detergent complexes. However, Jones proposes only a two-step interaction between cytoplasmic proteins (not membrane-bound proteins) and detergents: 1) detergent accumulates on binding sites on the protein up to saturation, and 2) additional detergent denatures the protein. The differences between the detergent-membrane and detergent-protein interactions suggest that more detergent may be needed to interact with the membranes than with the proteins. The RBC membrane composition is reported to be 19.5% (w/w) water, 39.5% protein, 35.1% lipid and 5.8% carbohydrates [42]. Therefore, a large increase in the amount of RBC membrane present in a mixture would then require a significant increase in the amount of detergent needed to solubilize all of the RBC membrane sufficiently for filtration, especially if the proteins need to be removed from the protein-lipid-detergent complexes formed by solubilization.
A main difference between filtration of whole blood and filtration of plasma recovered from the spinning disk process is the number of RBCs in suspension. Our spinning disk technology removes approximately 99% of the RBCs from the plasma in about 2 minutes, essentially dropping the concentration of RBCs by 2 orders of magnitude. The results of Table 1 show that this large decrease in total lipids and membrane-bound proteins requires a significant decrease in the amount of detergent needed to produce successful filtration of the same amount of spun plasma as is required to successfully filter whole blood.
However, Figure 1 highlights the fact that detergents can lyse or compromise the viability of bacteria. Thus, excessive detergent concentrations can lyse the bacteria in blood, while insufficient detergent concentrations may inadequately disrupt the blood cell membranes, allowing them to block the filter. Comparing the lysis and filtering results of Table 1 with the bactericidal results of Figure 1 shows that the necessary SDS concentrations to keep bacteria viable cannot be obtained for whole blood samples at small dilution volumes, but may be achieved for the spun plasma samples which contain much less blood cells. We therefore propose that there is a critical detergent-to-membrane ratio which specifies the minimum amount of detergent needed to sufficiently solubilize a given amount of membrane. Then by determining the maximum detergent concentration that will not kill bacteria, the maximum concentration of membrane allowable in a sample can be calculated.
Using a correlation between membrane concentration and Hct of the blood, the necessary dilution of the blood could be calculated. This would allow clinics to simply measure the Hct of the patient’s blood and then diluted according to a formula based on the critical detergent-to-membrane ratio. While possible, such a calculation could lead to impractically large dilution volumes if the bacteria are more sensitive to the surfactants employed.
The data of Table 2 reveals how proteases can reduce the dilution volume needed to achieve sufficient solubilization for successful filtration. For example, adding 33 mg of the proteinase from Aspergillus melleus in the lysis solution changed instant clogging of the blood-lysis-solution blend to complete passage of the mixture. The addition of a protease drastically increased the filtered volume of the whole blood. A similar filtration improvement was achieved using the spun plasma, instead of whole blood. The proteases, which cleave proteins, may facilitate protein removal from the cell membranes, which would decrease the amount of detergent needed for sufficient membrane solubilization.
Another potential problem with surfactant-facilitated whole blood filtration is the propensity of detergents to make stable complexes with the released proteins and cell membrane fragments [43]. Depending on the concentration and amount of unbound detergent in solution, these complexes can form precipitates that could potentially clog the filter [44]. However, chaotropes have been shown to weaken protein-detergent interactions and increase the solubility of the complexes [21,45]. In addition to weakening protein-detergent interactions, chaotropes also disrupt cell membranes, [46,47] which has both positive and negative outcomes – positive in that less detergent may be needed with the addition of the chaotrope, but negative in that chaotropes may increase the potential of lysing the bacteria.
Tables 3 & 4 reveal that the addition of salts to the blood lysis solution influences the filterability of the blood-lysis blend and is sensitive to the detergent concentration, salt concentration, and salt chemistry. The salts from Table 3 with which 100% of the blood- lysis blends were filtered comprise calcium chloride, lithium chloride, manganese chloride, potassium chloride, sodium chloride, potassium carbonate, potassium hydroxide, sodium carbonate, and sodium hydroxide. According to the Hofmeister series, the anions usually affect the system more than the cations. However, chloride is a fairly neutral anion on the kosmotropic-chaotropic scale while sodium is a weakly kosmotropic cation and potassium a weakly chaotropic cation. Thus, most of the salts that produced 100% filtration of the blood-lysis blend were relatively neutral salts. Yet, manganese chloride, potassium carbonate, and sodium carbonate are stronger kosmotropic salts. It is noteworthy that these stronger kosmotropic salts produced 100% filtration while mid-range kosmotropic salts such as sodium bicarbonate and sodium acetate did not. It is also important to note that none of the salts produced 100% filtration at a 1 M concentration for any of the 3 concentrations of Triton X-100. This implies that there is a combination effect present which cannot be adequately described by analyzing the phenomena produced by the detergent or the salts individually.
Table 4, which analyzes different salt concentrations for a given detergent concentration, supports the previous claim of a combination effect since the concentrations of the salts affect the filterability of the blood-lysis blend. Table 5 explored the combination effects for 3 types of sodium salts (sodium acetate, sodium bicarbonate, and sodium chloride) by analyzing different salt and detergent concentration combinations. For each of the 3 salts there were at least 2 different combinations of salt and detergent that produced 100% filterability of the blood-lysis blend, showing that both sodium bicarbonate and sodium acetate, which are mid-range kosmotropic salts, could produce 100% filtration of the blood-lysis blend. For sodium chloride, there were 9 different combinations of salt and detergent that produced 100% filtration of the blood-lysis blend, indicating this more neutral salt to be more versatile.
Tables 6 and 7 show that the number of blood cells present affects the ability of the combination of the salt and detergent to achieve lysis and 100% filtration of the blood-lysis blend. Table 6, which evaluated at a high Hct the effectiveness of the successful salt-detergent combinations from Table 5, reveals the need for developing a metric based on the Hct of the blood. Table 7 further explored the effects of the salt and detergent concentrations at different Hct. These results revealed that filterability was more sensitive to the salt concentration than to the detergent concentration. In fact, some of the detergent-salt concentrations used may not have been sufficient to lyse all the blood cells at high hematocrit.
To summarize the above discussion, filtration of blood cells depends on many factors, some of which are the hematocrit, the salt chemistry and concentration, and the detergent chemistry and concentration. Temperature and pH may also play a role but were beyond the scope of this study. The best filtration procedure found for spun plasma employed a 2% saponin solution with 33 mg protease, which allowed filtration of 4 mL of spun plasma at a 1:1 ratio (plasma:solution). The best lysis solution for whole blood employed a 4% Triton X-100 solution with 0.5 M NaCl which allowed filtration of whole blood at a 1:9 ratio (blood: lysis solution). Despite these encouraging findings, further research should be done to explore other chemistries that might better enable filtration of diluted whole blood.
Backflushing bacteria from filters
There are many factors which affect bacterial adhesion to filter surfaces, such as bacterial concentration, time of exposure, fluid shear stress, temperature, surface chemistry of both filter and bacteria, pH, ionic strength of solution, presence of proteins or other molecules (such as lipids and membranes), and roughness of the filter surface [48,49]. The suspension containing the bacteria and various lytic agents (filtration blend) consists of many proteins and lipids from the lysed blood cells as well as detergents and salts. Along with the bacteria, each of these substrates interacts with the polymeric surface of the filter, most probably modifying the surface characteristics. With so many variables present, it is difficult to distinguish the major factors in preventing or promoting the removal of the bacteria from the filter. However, the results shown in Figure 2 lead to the conclusion that the proteins and lipids from the lysed blood cells are a key component in promoting bacterial adherence to the filter. The details of how these molecules are affecting the bacteria and/or filter surface cannot be determined from the experiments performed, but the results of Figures 4B & 5B show that prior interaction of some detergents with the filter (pre-soaking the filter) may lessen some of the protein and lipid interactions that promote bacterial adherence. This is consistent with other studies [14].
An intriguing observation is that, while in general, bacteria are often reported to adhere better to hydrophobic surfaces than hydrophilic surfaces [50] our data in Figures 4 & 5 suggest that greater bacterial removal was attained using the hydrophobic filters. This data implies a simplistic postulate that the biomolecules in solution or on the bacteria surface interact more with the PVP polymer coating the surface of the hydrophilic filters than with the bare polycarbonate surface of the hydrophobic filters. A published study on plasma proteins and their effects on bacterial adherence to polycarbonate surfaces showed that the proteins decreased adherence to the surface while increasing bacterial surface charge [51]. However, a different study on protein-coated polymer surfaces showed that surfaces containing proteins in general had greater bacterial adherence [52]. Also, as the time of contact between the bacteria and the surface increased, bacterial adherence to the surface increased, but increases in ionic strength decreased bacterial adherence to the surface. Another study analyzed the difference between reversible and irreversible adherence to glass surfaces and found that high ionic strength was associated with irreversible adherence [53].
Still other studies have shown that polymeric substances, lipopolysaccharides, and proteins in the outer membrane of bacteria increase adhesion to surfaces [54-56]. Thus, the partial solubilization of the bacterial outer membrane may be advantageous in the removal of bacteria from the filter, as long as the bacteria are not lysed.
The complex biochemistry of lysed blood make it difficult to discern what exactly is occurring on the surface of the filters and all the interactions that reduce the bacterial recovery by backflushing. Thus we rely on our extensive experimental results that show a few important general trends. First, non-PVP-coated polycarbonate filters tend to release bacteria better than PVP-coated filters. Second, pre-soaking the filters with surfactant before bacterial deposition appears to aid subsequent removal during backflushing. Of the several backflush solutions examined, Tween-80 appears to be more successful in bacterial removal than Brij 58, Pluronic F108 and Tween-20, in that order; but combinations appear more effective. To summarize the above discussion, removal of bacteria from filter surfaces depends on many of the same factors as the filtration of lysed blood cells. But the trends for each step do not always trend in the same direction. However, we were able to achieve greater than 50% bacterial recovery under several different solution combinations for filtration and removal, with the optimum combination being incubation in a 1% Brij 58, 1% Tween-80, and 1% Pluronic F108 with 0.5M NaCl solution, followed filtration on a pre-soaked hydrophobic filter, and then backflushing with a 3% Pluronic F108. These findings provide great insight into the search for an optimum solution combination for removal of bacteria from blood followed by removal of the bacteria from the filter.
The combination of filtration and backflushing require only about 3 minutes in total, when using filtration solutions which sufficiently lyse the blood cells so that the filter does not clog. When the subsequent downstream process may not accommodate the chemicals required for whole blood lysis (4% Triton X-100 in 0.5 M NaCl), a slightly longer process time would be required to remove the majority of blood cells by spinning in a centrifugal disk for 3 minutes, followed by lysis with saponin and protease. The reagents used in backflushing also need to be considered for compatibility with downstream procedures. In any event, these front-end procedures to separate bacteria from whole blood are much more rapid than conventional growth methods for determining bacterial species in blood infections. For example this rapid isolation can be followed by PCR techniques to quickly identify resistance genes [57] or other critical genes in the invasive bacteria in sepsis. Such rapid identification will lead to more rapid personalized diagnostics to treat blood infections in a timely and directed manner.
Data Availability
Data are freely available by contacting the corresponding author.
Acknowledgements
Funding for this research came from the NIAID under grant R01AI116989 and from the Chemical Engineering Department of Brigham Young University.
Conflict of Interest
There are no conflicts of interest related to this research or this publication.
References
- Karam G, Chastre J, Wilcox MH, Vincent JL (2016) Antibiotic strategies in the era of multidrug resistance. Crit Care 20(1): 136.
- Yagupsky P, Nolte FS (1990) Quantitative aspects of septicemia. Clin Microbiol Rev 3(3): 269-279.
- Leggieri N, Rida A, François P, Schrenzel J (2010) Molecular diagnosis of bloodstream infections: planning to (physically) reach the bedside. Curr Opin Infect Dis 23: 311-319.
- Kumar A, Roberts D, Wood KE., Light B, Parrillo JE, et al. (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34(6): 1589-1596.
- Schweizer ML, Furuno JP, Harris AD, JOHNSON JK, Shardell MD, et al. (2011) Comparative effectiveness of nafcillin or cefazolin versus vancomycin in methicillin-susceptible Staphylococcus aureus bacteremia. BMC Infect Dis 11: 279.
- Opota O, Jaton K, Greub G (2015) Microbial diagnosis of bloodstream infection: towards molecular diagnosis directly from blood. Clin Microbiol Infect 21(4): 323-331.
- Diekema DJ, Pfaller MA (2013) Rapid detection of antibiotic-resistant organism carriage for infection prevention. Clin Infect Dis 56(11): 1614-1620.
- Vergara-López S, Domínguez M, Conejo M, Pascual Á, Rodríguez-Baño J (2014) Lessons from an outbreak of metallo-β-lactamase-producing Klebsiella oxytoca in an intensive care unit: the importance of time at risk and combination therapy. J Hospital Infect 89(2): 123-131.
- Duerden B, Fry C, Johnson AP, Lcox MH (2015) The Control of Methicillin-Resistant Staphylococcus aureus Blood Stream Infections in England. Open Forum Infect Dis 2(2): ofv035.
- Pitt WG, Alizadeh M, Blanco R, Hunter AK, Bledsoe CG, et al. (2019) Factors affecting sedimentational separation of bacteria from blood. Biotechnol Prog 36(1): e2892.
- Winn WR, White ML, Carter WT, Miller AB, Finegold SM (1966) Rapid diagnosis of bacteremia with quantitative differential-membrane filtration culture. JAMA 197(7): 539-548.
- Finegold SM, White ML, Ziment I, Winn WR (1969) Rapid diagnosis of bacteremia. Appl Microbiol 18(3): 458-463.
- Farmer SG, Komorowski RA (1972) Evaluation of the Sterifil lysis-filtration blood culture system. Appl Microbiol 23(3): 500-504.
- Zierdt CH, Kagan RL, Maclowry JD (1977) Development of a lysis-filtration blood culture technique. J Clin Microbiol 5: 46-50.
- Heimdahl A, Josefsson K, Von Konow L, Nord CE (1985) Detection of anaerobic bacteria in blood cultures by lysis filtration. Eur J Clin Microbiol 4(4): 404-407.
- Zierdt CH (1986) Simplified lysed-blood culture technique. J Clin Microbiol 23(3): 452-455.
- Zierdt CH (1982) Blood-lysing solution nontoxic to pathogenic bacteria. J Clin Microbiol 15(1): 172-174.
- Tang L, Huynh KA, Fleming ML, Larronde-Larretche M, Chen KL (2015) Imparting antimicrobial and anti-adhesive properties to polysulfone membranes through modification with silver nanoparticles and polyelectrolyte multilayers. J Colloid Interface Sci 451: 125-133.
- Zierdt CH (1979) Adherence of bacteria, yeast, blood cells, and latex spheres to large-porosity membrane filters. Appl Environ Microbiol 38(6): 1166-1172.
- Ball P, Hallsworth JE (2015) Water structure and chaotropicity: their uses, abuses and biological implications. Physical Chemistry Chemical Physics 17: 8297-8305.
- Moelbert S, Normand B, De Los Rios P (2004) Kosmotropes and chaotropes: modelling preferential exclusion, binding and aggregate stability. Biophys Chem 112(1): 45-57.
- Sammalkorpi M, Karttunen M, Haataja M (2009) Ionic surfactant aggregates in saline solutions: sodium dodecyl sulfate (SDS) in the presence of excess sodium chloride (NaCl) or calcium chloride (CaCl(2)). J Phys Chem B 113(17): 5863-5870.
- Arakawa T, Timasheff SN (1982) Preferential interactions of proteins with salts in concentrated solutions. Biochemistry 21(25): 6545-6552.
- Arakawa T, Timasheff SN (1984) Mechanism of Protein Salting in and Salting out by Divalent-Cation Salts - Balance between Hydration and Salt Binding. Biochemistry 23(25): 5912-5923.
- Arakawa T, Bhat R, Timasheff SN (1990) Preferential interactions determine protein solubility in three-component solutions: the MgCl2 system. Biochemistry 29(7): 1914-1923.
- Timasheff SN (1998) Control of protein stability and reactions by weakly interacting cosolvents: the simplicity of the complicated. Adv Protein Chem 51: 355-432.
- Robinson DR, Jencks WP (1965) The Effect of Concentrated Salt Solutions on the Activity Coefficient of Acetyltetraglycine Ethyl Ester. J Am Chem Soc 87: 2470-2479.
- Kragh-Hansen U, Le Maire M, Moller JV (1998) The mechanism of detergent solubilization of liposomes and protein-containing membranes. Biophysical Journal 75(6): 2932-2946.
- Levitzki A (1985) Reconstitution of Membrane-Receptor Systems. Biochimica Et Biophysica Acta 822(1): 127-153.
- Prete PS, Malheiros SV, Meirelles NC, De Paula E (2002b) Quantitative assessment of human erythrocyte membrane solubilization by Triton X-100. Biophys Chem 97(1): 1-5.
- Casadei BR, Domingues CC, Clop EM, Couto VM, Perillo MA, et al. (2018) Molecular features of nonionic detergents involved in the binding kinetics and solubilization efficiency, as studied in model (Langmuir films) and biological (Erythrocytes) membranes. Colloids Surf B-Biointerfaces 166: 152-160.
- Prete PS, Gomes K, Malheiros SV, Meirelles NC, De Paula E (2002a) Solubilization of human erythrocyte membranes by non-ionic surfactants of the polyoxyethylene alkyl ethers series Biophys Chem 97(1): 45-54.
- Pitt WG, Alizadeh M, Husseini GA, Mcclellan DS, Buchanan CM, et al. (2016) Rapid separation of bacteria from blood-review and outlook. Biotechnol Prog 32(4): 823-839.
- Alizadeh M, Wood RL, Buchanan CM, Bledsoe CG, Wood ME, et al. (2017) Rapid separation of bacteria from blood - Chemical aspects. Colloids Surf B Biointerfaces 154: 365-372.
- Buchanan CM, Wood RL, Hoj TR, Alizadeh M, Bledsoe CG, et al. (2017) Rapid separation of very low concentrations of bacteria from blood. J Microbiol Methods 139: 48-53.
- Wood RL, Whitehead JP, Hunter AK, Mcclellan DS, Pitt WG (2019) An experimental investigation of interfacial instability in separated blood. Aiche Journal 65: 1376-1386.
- Anderson CM, Pitt WG (2020) Effect of dilution on sedimentational separation of bacteria from blood. Biotechnol Prog 36(6): e3056.
- Sullivan NM, Sutter VL, Finegold SM (1975) Practical aerobic membrane filtration blood culture technique: development of procedure. J Clin Microbiol 1: 30-36.
- Costill DL, Saltin B (1974) Changes in the ratio of venous to body hematocrit following dehydration. J Appl Physiol 36(5): 608-610.
- Horino T, Chiba A, Kawano S, Kato T, Sato F, et al. (2012) Clinical characteristics and risk factors for mortality in patients with bacteremia caused by Pseudomonas aeruginosa. Intern Med 51(1): 59-64.
- Jones M (1992) Surfactant interactions with bio membranes and proteins. Chemical Society Reviews 21: 127-136.
- De Oliveira S, Saldanha C (2010) An overview about erythrocyte membrane. Clin Hemorheol Microcirc 44(1): 63-74.
- Putnam FW (1948) The Interactions of Proteins and Synthetic Detergents. Advances in Protein Chemistry 4: 79-122.
- Putnam FW, Neurath H (1944) The Precipitation of Proteins by Synthetic Detergents1a. Journal of the American Chemical Society 66: 692-697.
- Rabilloud T, Adessi C, Giraudel A, Lunardi J (1997) Improvement of the solubilization of proteins in two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 18(3-4): 307-316.
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-Van Dillen PM, et al. (1990) Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28(3): 495-503.
- Mahalanabis M, Al-Muayad H, Kulinski MD, Altman D, Klapperich, C. M. 2009. Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip, 9, 2811-7.
- Nnadozie CF, Lin J, Govinden R (2015) Selective isolation of bacteria for metagenomic analysis: Impact of membrane characteristics on bacterial filterability. Biotechnol Prog 31(4): 853-866.
- Katsikogianni M, Missirlis YF (2004) Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur Cell Mater 8: 37-57.
- Kochkodan VM, Hilal N, Goncharuk VV, Al-Khatib L, Levadna TI (2006) Effect of the surface modification of polymer membranes on their microbiological fouling. Colloid Journal 68: 267-273.
- Carballo J, Ferreiros CM, Criado MT (1991) Influence of Blood Proteins in the Invitro Adhesion of Staphylococcus-Epidermidis to Teflon, Polycarbonate, Polyethylene and Bovine Pericardium. Revista Espanola De Fisiologia 47(4): 201-208.
- Xu LC, Logan BE (2006) Interaction forces measured using AFM between colloids and surfaces coated with both dextran and protein. Langmuir 22(10): 4720-4727.
- Vigeant MAS, Ford RM, Wagner M, Tamm LK (2002) Reversible and irreversible adhesion of motile Escherichia coli cells analyzed by total internal reflection aqueous fluorescence microscopy. Appl Environ Microbiol 68(6): 2794-2801.
- Razatos A, Ong YL, Sharma MM, Georgiou G (1998) Evaluating the interaction of bacteria with biomaterials using atomic force microscopy. Journal of Biomaterials Science-Polymer Edition 9(12): 1361-1373.
- Fang HHP, Chan KY, Xu LC (2000) Quantification of bacterial adhesion forces using atomic force microscopy (AFM). journal of microbiological methods 40: 89-97.
- Burks GA, Velegol SB, Paramonova E, Lindenmuth BE, Feick JD, et al. (2003) Macroscopic and nanoscale measurements of the adhesion of bacteria with varying outer layer surface composition. Langmuir 19: 2366-2371.
- Hoj TR, Mcneely B, Webber K, Welling E, Pitt WG, et al. (2021) A pentaplex real-time PCR assay for rapid identification of major beta-lactamase genes KPC, NDM, CTX, CMY, and OXA-48 directly from bacteria in blood. J Med Microbiol 70(12): 001465.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.