Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Biological Factors and its Effect on HSP70 in Corbicula fluminea
*Corresponding author: Ashwin Sidhu, California Northstate University College of Medicine, 9700 W. Taron Dr., Elk Grove, CA, USA.
Received: August 07, 2023; Published: August 14, 2023
DOI: 10.34297/AJBSR.2023.19.002636
Abstract
Research was conducted on the clam species, Corbicula fluminea. This research is significant because the Asian clam is an invasive species, and it is essential to understand how biological factors encourage its population growth. The project focuses on HSP70 in the clams and the water and soil of the American River. These qualities are crucial to this project because they impact the survival of the clams. Overall, invertebrate feeding, microbial analysis of water and soil, and HSP70 expression was examined. The hypothesis tested is, if the water and soil conditions are not ideal, then the HSP70 protein levels will be elevated in the mantle samples. By conducting tests on water quality, it was concluded that clams use filtration to feed on microorganisms in the river, such as algae or bacteria. HSP70 expression was studied using SDS-PAGE and Western Blotting technique as well as Image J to closely observe the expression. Furthermore, testing for microbes in the water and soil, using coli plate techniques and agar plating, allowed for conclusions to be made on the stress levels of the clam species and its relation to HSP70. From experimentation, water quality was normal, which led to very faint bands of HSP70 due to little stress on the clam species. Normal water conditions, with some bacteria, allow Corbicula fluminea to be an invasive species in the American River.
Keywords: Corbicula fluminea, Invasive species, American river, HSP70 expression, Heat shock protein
Introduction
The Asian clam is considered an invasive species and is a problem in many freshwater ecosystems around the world. The Asian clam has spread throughout the world at an alarming rate, especially since the Asian clam was originally found in just Asiatic ecosystems. Specifically, the Asian clam species, also known as Corbicula fluminea, has spread throughout the world over the last century and has become a major invasive species in North America and Europe because it is considered a threat to the native aquatic communities [1]. The effects and importance of the Asian clam in lakes and waterways has been frequently debated and there are numerous positions that view the invasive species as detrimental to the environment. The Asian clam has made its way to the American River of Lake Natoma and is demonstrating its invasive capabilities. The American River is a home to various wildlife and fish and the Asian clam has a widespread presence in the river. The clam species, indigenous to Eastern Asia, finds itself competing with native species for space and nutrients in the river. It is important to understand that invasive species cause imbalances within the environment and they can cause many problems for native plant life and the ecosystem [2].
Different variables from the American River will be tested to help explain why the Asian clam, or Corbicula fluminea, is able to be so successful in the river. It is difficult to pinpoint the exact reason why the clam species is thriving in the American River, but experimentation will be done to help explain this question. The hypothesis to be tested is, if the water and soil conditions are ideal in the American River, then the HSP70 protein levels will not be elevated in the mantle samples, which will mean a successful invasive species due to no environmental stressors on the clam.
To test the hypothesis, it is important to understand the habitat and physical description of the Asian clam, such as the presence of the HSP70 protein. The habitat of the Asian clam is lake and rivers. Particularly, the Asian clam thrives in fast flowing water since the water is then able to provide food to the clam species. Also, the Asian clam is ectothermic, meaning the clam is dependent on external sources for body heat, and has bilateral symmetry. Also, the Asian clam has a high population density, which makes space very important. Lakes and streams give the Asian clams plenty of space to grow and reproduce [3]. In relation to food, clams are filter-feeders. This means that the primary food source for clams is small organisms, such as algae or bacteria. The American River may have algae and bacteria that acts as the food source of the clams, and testing will be done to confirm this notion. Importantly, the Asian clams have a protein, known as the HSP70, that are great markers of cellular stress. The heat shock protein, HSP70, will be studied because they are essential parts of the cellular network of molecular chaperones and contribute to catalyst folding [4]. Typically, if there are environmental stressors on a species, the level of HSP70 expression will be elevated. By studying the protein expression levels of HSP70, we will get a better understanding for how the Asian clam is able to successfully survive as an invasive species.
To conduct the testing and analysis of HSP70, the SDS-PAGE Western Blotting technique will be used, as well as computer software known as ImageJ. These techniques will help show the amount of HSP70 detected in various samples and the techniques will help provide insight and specific data on the protein of interest (HSP70). It is known that the survival of the Asian clams depends on specific factors. Specifically, the clam can survive in water that meets its needs, such as sufficient food availability. To test for bacteria in the water and soil, coli plate techniques and agar plating will be used. Furthermore, to get a better understanding of the feeding rates in Corbicula fluminea, the clams will be placed in a beaker with microalgal marine food and E. coli, to observe where the most feeding occurs. a certain concentration of nitrate and phosphate and an optimal pH and temperature of the water. If the Asian clams has some sort of stress placed on it, then the levels of HSP70 expression will be elevated and the HSP70 protein can be closely examined with Image J. It is important to realize the level of the HSP70 protein in a cell will vary depending on the stress level and the amount of time the stressful conditions lasted [5]. Overall, is necessary to know how much bacteria are present in the water and what types of bacteria there are, so students will have a better understanding of what the clams feed on. This will help with the connection between water and soil qualities and the expression of HSP70 in the Asian clams.
Procedure and Methods
Collection and Dissection
Field Collection: The purpose of this experiment was to collect the clams, specifically the Corbicula fluminea, that was needed for this experiment. To do the field collection, the waterproof waders were put on. The shovel was used to dig and bring dirt up as well as clams. This process was repeated until enough clams had been collected and placed in a container. After the clams were collected, containers were used to obtain soil samples and water samples. Lastly, the temperature of the lake water was taken using the thermometer. In total, 11 clams were collected, and the water temperature was 68℃.
Dissections: One clam was taken out from the container which had many clams in it. Using the dissection tools, a gill sample and mantle sample from the clam was obtained. Immediately after obtaining the samples, the samples were placed in a small tube and put in ice. This process continued until three total clams were dissected. In other words, three samples of mantles and three samples of gills were obtained.
Invertebrate Feeding
Observing Feeding Rates in Clams: The experiment helped measure filtration rates in clams. Also, bacterial clearance was observed. The feeding rate of the clams was observed in microalgal food. The clam was weighed, and the beaker was filled with water. A 96 well plate was used throughout this experiment to determine the absorbance and conclude whether feeding took place. Three wells were filled every 15 minutes. Also, in this experiment, the E. Coli was used as the food source for the clams. First, the clam was weighed on a weigh boat. Next, a E. Coli strain was obtained, and the cap was tightened on the vial. The E. Coli was prepared. The E. Coli broth was added to the water in the beaker. Again, a 96 well plate was used, and 3 readings were taken every 15 minutes for 2 hours.
Data Analysis: The feeding rate of the clams was smallest in the E. coli broth than with the super concentrated marine microalgal food. In other foods, the clam fed less in the E. coli broth. It was concluded that in the E. coli broth, the feeding rate was 4.0x10-5. This value was a lot smaller than the feeding rate obtained by the microalgal food. The feeding rate was represented by a linear equation for both microalgal food and the E. coli broth.
Cirri Spacing
Dissection: The purpose of the dissection was to prepare for cirri spacing analysis. Gill samples were needed, to examine the cirri spacing in the clams. To do the dissecting, a clam was placed in a dissecting tray and the anterior and posterior ends of the clam were identified. Also, the dorsal, ventral, and lateral surfaces were identified. The umbo, a bump at the posterior end of the valve, was located and a scalpel was placed on the umbo to break through the shell. Once the scalpel broke through the shell, the clam was rotated so the adductor muscles were cut cleanly. This opened the clam. Using the scissors, a sample of the gills were carefully cut off. The sample of the gills were placed on a mini glass plate, which were observed under a microscope.
Cirri Spacing: The cirri spacing was measured within the clams’ gills using the image and a software (Image J). Measurements were taken for three cirri in each clam to ensure accurate data was obtained.
Data Analysis: It was seen that the cirri spacing was different for each clam. Nonetheless, all the cirri spacing averages were between 0.02mm and 0.05mm. Again, from the cirri spacing it can be concluded that each clam has different feeding rates based on the length of the cirri spacing.
Microbial Analysis for Water and Soil Samples
Water Analysis: The purpose of the experiment was to see the different types of bacteria that are in the lake water and what the clams feed on. To conduct the water testing, the coli plate lid was removed and the water sample in the container was poured gently into the plate. As the pouring was occurring, it was ensured that the stream of water was entered into each well. When all the wells were full, the side of the coli plate was gently tapped to make sure that there were no air bubbles. Also, to make sure that there was no cross-contamination among the wells, any excess water was poured out. The lid was placed back on the coli plate. The water sample analysis was set up and the coli plate was placed in an incubator.
Soil Analysis: The purpose of this experiment was to plate diluted samples of the soil from the lake water and examine the bacteria that grows. Eppendorf tubes were labeled as 100 (1mL of soil water), 10-1 (900μL sterile water), 10-2 (900μL sterile water). The soil sample was mixed, and 1mL was added to tube 1 (100) using sterile tips. Tube 1 was mixed again and then 100μL was transferred to tube 2 (10-1). Then, tube 2 was mixed and then 100μL was transferred to tube 3 (10-2). After, three agar plates were labeled as: 10-1, 10-2, 10-3. When the agar plates were labeled, 100 μL from tube 1 was spread onto the 10-1 agar plate using a sterile swab. This process was repeated for tube 2 and 3 to the 10-2 and 10-3 agar plates respectively. The plates were inverted and incubated.
Data Analysis: For water analysis, the MPN, or most probable number of colony forming units per 100 mL sample, was found. 42 wells were blue, which meant the MPN was 146. This was determined using a chart. The number of wells which were “shining” under the UV light were counted. 1 well was glowing under UV light, which meant the MPN was 3. For soil analysis, calculations were done to determine the number of colony forming units. The colony forming units were 97402.597 CFU/g. soil.
Species Diversity and DNA Barcoding
DNA Extraction and Cell Lysis: The gill samples were collected, and cell lysis and DNA extraction were done. This was done by adding lysis buffer and Protease K to the sample tubes. Throughout the experiment, the centrifuge was used at 14000rpm. During the experiment, Silica Resin was added to the tubes and the supernatant was removed for many of the steps. The tubes were placed in a heat block and eventually a pellet containing genomic DNA was obtained. It was important to not disturb the pellet while doing the experiment.
Polymerase Chain Reaction and the COI Gene: The purpose of this experiment was to understand the process of the PCR and how it can be applied to amplify and detect specific target DNA sequences. Also, it was learned how to avoid contamination of DNA samples using clean lab techniques. To do this experiment, micro-centrifuge tubes were needed, and a master mix was created with deionized water, PCR buffer, primer mix, dNTPs, and Taq Polymerase. Once the master mix was completed, four PCR tubes were needed. Two were needed for the actual samples and two were needed for positive control and negative control. After the PCR tubes were set, they were placed in the Thermocycler, and it was turned on.
Agarose Gel Electrophoresis of PCR Samples: The purpose of this experiment was to get familiar with agarose gel electrophoresis and be able to determine the sizes of DNA bands observed. Also, students understood how to interpret the PCR results and the process of DNA sequencing. To do the experiment, agarose gel was placed in a taped gel rig and allowed to cool until the gel solidified. The PCR reactions from the previous experiment were obtained and the samples were prepared with loading dye. The samples were inserted into the wells along with the DNA ladder using a micropipette. The power supply was turned on to 100V when the TBE buffer was added. After the gel was run, the gel was transferred to the UV trans-illuminator in the digital imager chamber. This allowed for the image of the gel and the movement of the DNA fragments to be shown. From this experiment, a semi-log graph was made that showed the distance migrated from the well and the size of the DNA fragments. From the graph, it was determined that the length of the COI gene, from our sample, was 600bps.
Bioinformatics: The purpose of this experiment was to get familiar with the concept of bioinformatics and study the DNA sequences of C. fluminea. In the experiment, BLAST was used to identify the target of the PCR primer. The internet browser allowed students to search the forwards and reverse primers for matches. After BLAST was familiarized, the software FinchTV was used to trim the sequence and view the sequence on the chromatogram. The files were saved as the experiment went on. After the sequence was trimmed in FinchTV, the sequence was inserted into BLAST to find organisms that matched significantly with the trimmed sequence. MEGA7 was then used to compare the trimmed sequence with the resulting sequence on BLAST. This allowed direct comparison of the sequences.
Protein (HSP70) Expression
Sample and Buffer Preparation: The purpose of this experiment was to prepare for SDS-PAGE and Western Blot analysis. In this experiment, total protein was extracted from C. fluminea mantle tissue. To prepare the samples, 500μL of buffer K was added per 100mg of tissue sample to the plastic tissue grinder. The tissue samples were grinded for 1minute on ice. The tissue samples were then transferred to 1.5mL Eppendorf tubes with a transfer pipet. This was done for all three samples of the mantle tissue. The protein- loading buffer, Beta-Mercaptoethanol (BME), was added to the tissue samples at a 1:1 ratio. The samples for boiled for 5 minutes and stored on ice until they were placed in the freezer.
SDS-PAGE and Western Blotting: The purpose of this experiment was to conduct SDS-PAGE and Western Blotting with the mantle tissue samples. The tissue samples were boiled on the heat block for 5minutes and then placed on the tube rack. The tetra cell and the power supply were set up. To do this, the gel was removed from the storage pouch and the tape was removed from the bottom of the gel. The gel was loaded into the transfer rig correctly and SDS-running buffer was added to the clamping frames. The lanes were loaded with the samples, as well as positive and negative controls. The gel was run after the power supply was set to 120V. To prepare for Western Blotting, 2 filter pads, 2 filter paper, PVDF membrane, and the gel was needed. A transfer sandwich/cassette was made and inserted into the rig with the black side of the cassette facing the black side of the rig. After Western Blotting was run, there was total protein staining of the membrane to assay for transfer efficiency. The membrane was stored.
Primary Antibody Incubation and Secondary Antibody Incubation: The purpose of this experiment was to set up for Western Blot analysis using antibodies. The membrane was blocked to prevent non-specific binding of the antibody. The membrane was placed in a small container and covered with BSA/TBST. A small heat sealable bag was used to incubate the blot in primary antibody. The diluted antibody solution was poured into the bag, and the last edge of the bag was heat sealed. The blot was incubated in primary antibody at room temperature on the rocker. The bag was opened, and the primary antibody was saved. The membrane was washed with TBST buffer a total of 3 times. The wash buffer was discarded down the drain and the secondary antibody was added to small container and put on the rocker. The blot was washed 3 more times with TBST. Eventually, chemiluminescent substrate was added to the small container to cover the blot. The membrane was imaged using a transilluminator, and the image was saved for later use.
Western Blot Analysis. The purpose of this experiment was to use ImageJ to examine the bands on the Western Blot. ImageJ was opened on the desktop. The image obtained from the imaging of the membrane was dragged into ImageJ. A rectangular area around the first band was selected. “CTRL” “1” was pressed to mark the box size. The cursor was moved so it sat over one of the box border and “CTRL” “2” was pressed to select the next band. The step was repeated until each subsequent band was selected. Once all the bands were selected, “CTRL” “3” was pressed to bring on the image which showed the histograms. The histograms indicated the intensity of each of the bands. A histogram was created for each of the bands. The first histogram was selected, and a line was drawn across the top of the histogram to calculate the “area.” The intensity of the band was given as a numerical value. In relation to this experiment, the brighter the band, the higher the number for intensity (Figure 1).

a) Algae Feeding Average and Standard Deviation: 0.036±0.005.
b) Bacteria Feeding Average and Standard Deviation: 0.035±0.003.
c) Cirri Spacing Average and Standard Deviation: 0.038±0.007.
Figure 1: Invertebrate Feeding: Panel A shows the average absorbance from algae feeding during clam feeding. The absolute value of the slope demonstrates the feeding rate of the clams. Panel B shows the average absorbance from bacteria feeding during clam feeding. Panel C shows the cirri spacing of a clam. Cirri spacing was examined after dissections of clams. Multiple clams were dissected and the average cirri spacing in millimeters was taken for each. The average cirri spacing is shown in panel D. The average cirri spacing was obtained by using the arrows and finding the length of the cirri. Each measurement was taken three times, and they were averaged to get the average cirri spacing in millimeters (mm).
Figure Narrative
Based on Figure 1, the feeding rate of the clams was smaller with the E. Coli broth than with the super concentrated marine microalgal food. In other words, the clam fed less in the E. Coli broth. From the absolute value of the slopes, it was determined that the feeding rate of the clams, on average, in the algae water was 0.0012. On the other hand, the feeding rate of the clams, on average, in the E. coli water was 0.0005. Thus, feeding rate is seen as larger in the algae water. The average and standard deviation for algae feeding was 0.036±0.005 and the average and standard deviation for bacteria or E. coli feeding was 0.035±0.003. From error analysis, it can be determined that it is possible that the clams fed more in the E. coli water. The standard deviation was the smallest with the E. coli feeding, so the clams fed more consistently in the bacteria containing water. Nonetheless, feeding occurred in both situations. As shown by panel D, image 23 had the largest average distance of cirri in mm. Also, it is shown that image 20 had the lowest average distance of cirri. This part of the project most likely had the most error because the measurements may not have been too accurate. Furthermore, the average and standard deviation of the all the cirri space images was 0.038±0.007. There is small deviation, so the all the clams dissected had cirri spacing close to the average. On average, the cirri spacing of the clams is 0.038millimeters. In addition, even though the measurements were done in Image J, it is possible that human error caused inaccurate measurements (Figure 2).

Figure 2: Microbial Analysis for Water and Soil Samples: Panel A shows the microbial analysis of the river water. Furthermore, panel A shows a 96 well plate after incubation testing for MPN (Most Probably Number) of E. Coli and coliform bacterium in the water sample from the American River. There is water in each well and there is no cross-contamination because it was made sure that the wells were not overflowing. After incubation, some of the wells turned blue. Depending on the color of the wells under the UV light, it was possible that the water contained E. Coli bacteria. Panel B-D shows that there are bacteria in the soil samples. The three agar plates were set up by diluting the soil sample which provides the colony forming units per gram of soil. All three agar plates had bacteria growth on it.
Figure Narrative
Figure 2 shows that there are bacteria in the soil samples. All three agar plates have bacteria growth on it. The agar plate labeled as 10-1 had the most bacteria growth. The agar plate labeled as 10-2 had the second most bacteria growth. Finally, the agar plate labeled as 10-3 had the least amount of bacteria growth. This is because the soil sample was getting diluted. Less and less amount of soil was being added to the Eppendorf tubes, so it would be easier to count the number of colonies. Panel D had 75 colonies of bacteria. 75 was used to make the calculation for colony forming unit. It was determined that the number of colony forming units in the soil sample was 97402.597CFU/g. soil. To get the number of colony forming units per grams, the dilution factor needed to be used as well (10- 3). It is a very tedious process to count the exact number of colony forming units in a sample. Thus, a simple calculation was done to estimate the number of colony forming units. Also, well plates were measured in MPN. MPN means the Most Probable Number of colony forming units per 100mL sample. The 42 blue wells translated to 146MPN. Also, the 1 shining blue well translated to 3 MPN (Figure 3).

Figure 3: Agarose Gel Electrophoresis to Determine Size of COI gene: Panel A shows the results of the agarose gel electrophoresis of the COI gene. To successfully conduct the agarose gel electrophoresis, a DNA ladder was needed, two samples from the PCR reaction were needed, and a positive and negative control were needed. In panel A, each well represents a different sample that was loaded into the well. These were used to compare to each other and see if the PCR reaction was successful. Panel B shows the results of the agarose gel electrophoresis in graph form. The DNA ladder was graphed, and the COI gene is the sample was estimated using the best fit line. From the DNA ladder, the size of the DNA fragments, in Base Pairs (bps), from the gel electrophoresis can be determined.
Figure Narrative
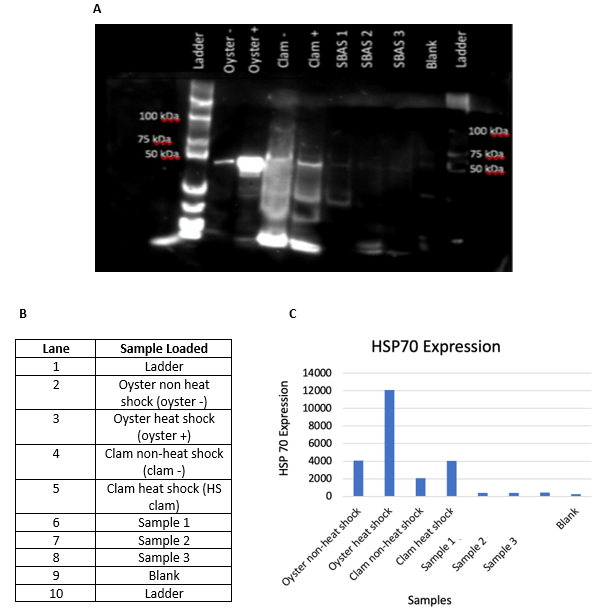
Figure 4: Protein (HSP70) Expression: Panel A shows HSP70 in the samples. To get the image in panel A, a transilluminator was used and the image was optimized, so it can be used later on to get the bar graph shown in panel C. In the blot image, there were three samples from the clams as well as positive and negative controls. As shown in panel B, ladder was loaded into lane 1, oyster non-heat shock was loaded into lane 2, oyster heat shock was loaded into lane 3, clam non-heat shock was loaded into lane 4, and clam heat shock was loaded into lane 5. Furthermore, three prepared mantle samples from C. fluminea were loaded into lanes 6, 7, and 8. Panel C was obtained from ImageJ as comparisons were made between the bands in different lanes.
The purpose of conducting agarose gel electrophoresis for the COI gene was to determine the sizes of the COI, or Cytochrome c oxidase subunit 1, in the sample and determine if it matched up with the typical size of the COI gene (~700 bps). In other words, the experiment was run to see if the PCR reactions were successful, and to see if the COI gene was successfully amplified for the succeeding experiment, DNA barcoding. The agarose gel was loaded with both a positive control and a negative control. The positive control used in the experiment was from flatworms. The agarose gel electrophoresis was run in a buffer at a 100 V. Based on the positive control, sample#2 showed the presence of the COI gene Figure 3. Sample#2 was focused on in relation to the succeeding part of the experiment, DNA barcoding. After agarose gel electrophoresis, sample#2 was found to have a COI gene with a size of 700 bps Figure 3. Sample#2 was chosen because it corresponded to the positive control. On the other hand, sample#1, while it was supposed to have a band matched up with the positive control, did not clearly show the appearance of the COI gene. Therefore, DNA barcoding was done on sample#2.
Once the results were returned, DNA barcoding was conducted, but the returned results were inconclusive because there were many unknown sequences. With too many unknown sequences, there was no way to accurately compare species diversity. The purpose of DNA barcoding in this experiment, was to compare species diversity with that of the Asian Clam. Nonetheless, there was no data to report from the DNA barcoding with the COI gene (Figure 4).
Figure Narrative
The purpose of SDS-PAGE and Western Blotting was to examine the HSP70 in the clam species. Also, the controls were important for this part to ensure that the protocol was successfully followed and there were no false positives obtained from the experimentation. The use of ImageJ was important in this part of the project in order to get visual results for the expression of the HSP70 in each of the loaded samples. Overall, ImageJ allowed for quantitative analysis to support that mantle samples tested had minimal HSP70 presence. As shown in panel A, the HSP70 expression was located around 70-75kDa. From panel C, it is seen that the oyster heat shock had the highest HSP70 expression, while all three mantle samples from the Asian clam had the lowest amount of HSP70 expression. Lane 9, which was supposed to be blank, showed minimal expression of HSP70. This means that there was some sort of contamination. It is possible that there was contamination from the other lanes which led to lane 9 being loaded with a sample, rather than it being left blank. From the HSP70 expression, conclusions were made connecting the relation between the bacteria found in the water and soil samples with the feeding rates, and further with the chemistry aspect of this project.
Discussion
By conducting microbial analysis on the water and soil samples from the American River, it was determined that there are bacteria in both the water and soil samples collected from the American River. This can be seen in the data/results section. Specifically, Figure 2 is evidence that there are bacteria present in the river water. Through coli plate techniques and agar plating, Figure 2 shows that many of the wells turned blue in the well plate after incubation. Based on just the blue colored wells, it can be swiftly determined that there are definitely coliforms in the river water. The way to determine if there are E. coli strains in the river water sample is by using the UV light and an MPN (most probable number) conversion sheet. Specifically, MPN means the most probable number of colony forming units per 100mL sample. The UV light was shined on the well plate in a dark room to see how many wells were shining bright. This indicated E. coli strains. Again, the 42 blue wells translated to 146MPN and the 1 shining blue well translated to 3 MPN. So, in the entire river water sample collected, there was 146 colony forming units of coliforms per 100mL of water. On the other hand, there was 3 colony forming units of E. coli per 100mL of water. Figure 2 also show that there are bacteria in the soil samples. By calculating the number of colony forming units in the soil, it was confirmed that the soil also has bacteria. This is an important development for this experiment because the clams have food to feed on. The number of colony forming units in the soil sample was 97402.597 CFU/g. soil. Thus, the water and soil samples both had bacteria in it.
There were a few limitations and some possible errors during the microbial analysis of the soil and water samples from the American River. The first major error that could have occurred was when the water was being poured in the well plate. It was important to not talk or breathe over the well plate because bacteria or germs could be transferred from my partner or I to the well plate. Another possible error could have been the inaccurate weighing of the soil sample that was used for the soil testing. For example, when the soil sample for the test was weighed, it was 0.77g. This lead to it being determined that the number of colony forming units in the soil sample was 97402.597 CFU/g. soil. If the soil was weighed to be heavier, then there would be less colony forming unit in the sample. Thus, inaccurate conclusions would have been made. Nonetheless, there is confidence that the errors were limited, and the conclusions made in the project were valid.
In comparison to the feeding rates experiment, it is probable that the bacteria in the water was the food for the clams. Clams are filter-feeders, so they take advantage of any microorganisms in the water. The results were not surprising because “Coliforms are a broad group of bacteria that can be found in soil, decaying vegetation, water and feces. Their presence does not always indicate a threat to health but could point to a problem with treatment operations or a breach in the distribution system” [6]. Thus, the number of coliforms and E. coli in the river water is not a problem to the overall health of the ecosystem and it provides the Asian clam with food that helps explain the results in the feeding rate part of the experiment.
From this research, it is evident that the feeding rate of the clam in the super concentrated microalgal food was a lot higher than the feeding rate of the clam in the E. Coli broth. This is known by looking at the equations on each graph. To find the filtration rate of the clams, the absolute value of the slope is looked at. Specifically, in panel A of Figure 1, the feeding rate was 0.0012. In comparison, panel B of Figure 1 represented the feeding rate of the clam in the E. coli broth, which shows the feeding rate to be 0.0005. Based on the slopes of the graphs in Figure 1, it can be concluded that the clams had a higher feeding rate with the microalgal food. The way that feeding took place in the clams is by the action of cirri. Specifically, the clam opens it shell not too wide and the water enters the shell cavity. Then, oxygen and food are brought into contact with the gills. As this is occurring, waste is removed. The capture of the food occurs because of the cirri. On each gill, there are rows of cirri [7]. The cirri move back and forth, and this action causes water to move through the gills and allows food to be captured. The cirri spacing differs, most likely based on clam size. Thus, clams have different feeding rates based on the length of the cirri spacing (panel D of Figure 1). For example, a clam with large cirri spacing length most likely feeds at a higher rate because the cirri are able to capture the food easier. This is important because it explains the differing feeding rates of C. fluminea.
It can be concluded that there is a correlation between the cirri spacing and the ability to filter food. Since algae is bigger in size it is easier for the cirri to trap the food particles. On the other hand, bacteria particles are not too big in size which would make it harder to filter the bacteria. However, bacteria presence in the water and soil is a big part of the ability of the clam to be an invasive species in the American River.
There were a few limitations in the feeding rate part of the research. One possible error was that the clam may not have fed properly if the lab space was too loud and if the table was moving too much. The clams may have gotten frightened, which decreased their feeding. If this is the case, then the feeding rates were not accurately observed. This would lead to inaccurate conclusions. Another possible error was during the cirri spacing measurements. It was important to make sure that the measurements were actually being taken of the cirri and not another part of the gill. This would have led to many errors during the construction panel C in Figure 1. Despite the possible error, there is confidence that the conclusions made from this research were accurate.
Figure 3 shows how the COI gene was studied using agarose gel electrophoresis and a semi-log graph for the DNA ladder. The COI gene is extremely important for this experiment because the “cytochrome c oxidase subunit I (COI) gene plays a pivotal role in a global effort to document biodiversity and continues to be a gene of choice in phylogenetic and phylogeographic studies” [8]. In part of the project, the graph for the DNA ladder was constructed in order to determine the size of the COI gene in the two samples resulting from the PCR reaction. The two PCR samples from the gill tissue were prepared and tested using agarose gel electrophoresis. Agarose gel electrophoresis was conducted in order to determine the size of the COI gene. This is important because the idea behind this technique is to separate the DNA fragments and compare them to positive and negative controls. The controls are needed anytime when conducting gel electrophoresis to ensure that the results are not due to chance. In other words, in the experiment, positive controls were used to indicate that the procedure is working, and the negative control is used to check for false positive results. The resulting gel is shown in panel A of Figure 3.
Gel electrophoresis was run on the PCR sample, in an attempt to conduct DNA barcoding on the clam species, Corbicula fluminea. DNA barcoding is a very useful method used by biologists to identify a species as belonging to a particular species. However, DNA barcoding could not be done in the experiment because there were too many unknowns in the sequencing return. Nonetheless, the agarose gel electrophoresis was successful because the positive control matched up with sample#2 (2 SBAS). This is clearly shown in panel A of Figure 3. From the gel electrophoresis it can confirmed that the COI gene is indeed present. This is important because the next step in the experiment was performed. The semi-log graph of DNA ladder (panel B of Figure 3) was made, and it showed that the size of the COI gene was, in fact, 700 base pairs. This is important because the presence of the COI gene was confirmed. While the gel electrophoresis showed the positive control corresponding to the sample PCR of gill tissue, the semi-log graph reinforced the presence of the gene. By understanding the COI gene and its presence in the clam, the biodiversity of the clam can be explored. There are numerous limitations with DNA barcoding. For example, this experiment exemplifies the limitations with this technique because if there are too many unknown base pairs, DNA barcoding cannot be completed. Also, DNA barcoding does not work as well when working with plants. Furthermore, “while DNA barcoding, using a single gene or standard set of genes may be appropriate for animals, it is not so for plants due to their dramatically different life history characteristics, evolutionary histories, and hybridization” [9]. This is why DNA barcoding can be done on animals and aquatic life, such as the Asian Clam. Overall, the concept and technique of DNA barcoding has numerous limitations.
Figure 4 shows the HSP70 expression in numerous samples, even though some samples showed very minimal HSP70 expression. HSP70 was studied throughout this experiment because “its level increases rapidly in response to various types of severe stress, as a protection against a subsequent, near-lethal, ischemic, or hypoxic event” [10]. Specifically, the oyster heat shock sample showed the most HSP70 expression. The reason for the high protein expression in the oyster was because the oyster was most likely highly stressed before tissue was extracted. On the other hand, it was determined that the Asian clams were not stressed because there were ideal conditions for their survival in the American River. Again, there was small levels of bacteria in the water and soil samples that were collected from the river. Since the Asian clam is a filter feeder, it is able to feed on small microorganism, such as bacteria and algae. In connection with the chemistry aspect of the experiment, there was both phosphate and nitrate in the water, which contributed to some levels of algae. So, both algae and bacteria (coliforms and E. coli) were present in the water and soil, which led to low expression of HSP70 in the clams. In other words, the clams were not stressed because they had a sufficient amount of food available to them. Based on panel A, the HSP70 bands can be seen around 75kDa. This is because the molecular weight of HSP70 is around 70kDa. Thus, panel A shows the HSP70 bands, while panel B shows where each sample was loaded. The ladders were used as the controls to compare the sizes of the bands in order to correctly identify HSP70. It made sense that the oyster also showed HSP70 expression because it is present in all organisms, from archaebacteria through plants to humans.10 Some limitations for the HSP70 part of the project was that SDS-PAGE and Western Blotting had to be done perfectly or it was possible that no protein would be shown. For example, the antibody incubation and substrate had to be done in a timely manner or the amplification of the protein will not be able to be observed under the transilluminator. Overall, all the experiments can be connected to conclude that HSP70 expression in Corbicula fluminea is limited because the clams are not stressed in the American River. They have the resources available to them to remain an invasive species in the American River.
Further research can be done on the Asian Clam and its ability to survive in the American River. For instance, the DNA barcoding part can be tried again to get enough readings to compare species diversity. More research can be done on different genes and even proteins. The relationship between the COI gene and the HSP70 protein can be examined. For instance, it can be studied if one effects the other and vice versa. Also, future research can be done on bacterial presence in the water and soil to observe changes over time. For instance, by testing the bacterial presence during different seasons, it is possible that the results obtained may be different. Furthermore, the HSP70 in the Asian clams can be tested during different seasons to see the difference in expression. If the levels stay in a relative range to which the Asian Clam is satisfied, then the growth of this invasive species will continue. If the trend shows a deviance from the ideal conditions, then the growth rate of the Asian Clam may decrease and eventually become negative. This would, in turn, lead to the possible elimination of Asian Clam from the American River. If more research is done on the clam species, Corbicula fluminea, it will be understood as to why the clam is an invasive species and why it is widespread in waterways around the world.
Conclusion
The purpose of this experiment was to conduct research to explain why the Asian clam is able to thrive as an invasive species in the American River. The overarching hypothesis connecting all of the experiments was, if the water conditions are not within the ideal range for the clam, then the HSP70 protein levels will be elevated in the mantle samples. In order to test this, various experiments were conducted.
Through experimentation, knowledge was gained on the Asian clam and the feeding habits of the species. A basic understanding of the food sources of clams and the process the clams go through to eat was obtained. Cirri can filter algae easier because the particles are a lot bigger than the particles of bacteria. From the feeding rate and cirri spacing part, it was concluded that the Asian clam takes advantage of the bacteria and algae that is in the waterways to be successful. Furthermore, from experimentation on the water and soil samples from the river water, it was concluded that there is both coliforms and E. coli in the water. Additionally, the COI gene was amplified through a PCR reaction, which allowed this experiment to be conducted. Specifically, this experiment allowed practice with agarose gel electrophoresis as well as creating a semi-log graph. A basic understanding about the COI gene in the gill tissue samples was obtained.
In relation to HSP70 in the project, the hypothesis was confirmed because there were very faint bands due to there being ideal conditions in the water. Also, HSP70 can be related back to the chemistry aspect of the project because the nitrate levels, phosphate levels, and dissolved oxygen levels are chemical factors that impact the survival of the Asian clam. Again, the levels were all ideal and within the normal range, so the clams are able to be successful invasive species. The most important outcome of the biology aspect of the project was that the clam species, Corbicula fluminea, is not stressed in the American River. This was confirmed from the HSP70 expression in the mantle samples. The Asian clam will continue to be an invasive species in the American River, unless there is a trend that shows an abnormality from the ideal conditions. Overall, the Asian clam is an invasive species in the American River because there are no environmental stressors that limit or obstruct its ability to spread throughout waterways.
Acknowledgement
Thank you to California Northstate University College of Health Sciences for providing the necessary materials as well as the mentorship in the completion of this research project.
Conflict of Interest
None.
References
- Chen H, Zha J, Liang X, Jihong B, Wang M, et al. (2013) Sequencing and De Novo Assembly of the Asian Clam (Corbicula fluminea) Transcriptome Using the Illumina GAIIx Method. PLoS One 8(11): e79516.
- Sousa R, Antunes C, Guilhermino L (2008) Ecology of the invasive Asian clam Corbicula fluminea in aquatic ecosystems: an overview. Int J Lim 44(2): 85-94.
- Denton ME, Chandra S, Wittmann ME, Reuter J, Baguley JG (2012) Reproduction and Population Structure of Corbicula fluminea in an Oligotrophic Subalpine Lake. Journal of Shellfish Research 31(1): 145-152.
- Mayer MP, Bukau B (2005) Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci 62(6): 670-684.
- Hradilova S, Havrdova M, Panacek A, Kvitek L, Zboril (2015) Hsp70 as an indicator of stress in the cells after contact with nanoparticles. J Phys Conf Ser: 1-7.
- Ellis K, Gowdy C, Jakomis N, Bernadette R, Thom C, et al. (2018) Understanding the costs of investigating coliform and coli detections during routine drinking water quality monitoring. Urban Water Journal 15(2): 101-108.
- Neufeld D, Yoder J (2011) The Role of Feeding Adaptations in Resource Competition between Invasive and Native Clams. Proceedings of the Assoication for Biology Laboratory Education 32: 78-87.
- Buhay JE (2009) COI-Like Sequences are becoming problematic in molecular systematic and DNA barcoding studies. Journal of Crustacean Biology 29(1): 96-110.
- Reynaud DH, Mishler BM, Brown PN (2015) The Capabilities and Limitations of DNA Barcoding of Botanical Dietary Supplements. AuthenTechnologies: 1-13.
- Zsigmond J, Gombos T, Förhécz Z, Pozsonyi Z, Karádi I, et al. (2013) Elevated extracellular HSP70 (HSPA1A) level as an independent prognostic marker of mortality in patients with heart failure. Cell Stress Chaperones 18(6): 809-813.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.