Review Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Avian Influenza Virus: A Review
*Corresponding author: Leonard Ranasinghe MD, PhD, Professor of Emergency Medicine and Director of Fourth-year Electives and Sub-Internships, California Northstate University College of Medicine, Elk Grove, CA
Received: September 05, 2023; Published: September 11, 2023
DOI: 10.34297/AJBSR.2023.20.002674
Abstract
There are four major types of the avian influenza virus: A, B, C and D. The H5N1 (Hemagglutinin type 5, neuraminidase type 1) strain of the avian influenza virus is part of the A subtypes and inflicts human viral. Once an individual is infected, either HPAI (Highly Pathogenic Avian Influenza) or MPAI (Mildly Pathogenic Avian Influenza) related symptoms could occur, depending on pre-existing diseases and risk factors. All subtype A influenza viruses contain eight single stranded negative sense RNA segments, each encoding for significant proteins that are utilized predominantly during the viral life cycle and replication process. The subtypes of the avian influenza virus arise from the 15HA (Hemagglutinin) and 9NA (Neuraminidase) protein variants. This review discusses how each protein of the virus plays a role in the pathogenicity of the virus, common clinical symptoms as well as an atypical case of infection, host cell immunology and which cell signaling pathways become interrupted upon infection, mechanisms of the Influenza Virus evolution, transmission of H5N1 to humans and within poultry and wild birds and treatment methods.
Introduction
Highly Pathogenic Avian Influenza (HPAI) is a viral disease that spreads through poultry, particularly chicken. With the rise of HPAI H5N1 on the rise, it is starting to become considered as a potentially serious pandemic threat. H5N1 infects both poultry and humans, particularly from infected poultry to humans. Of the 15 known types of HA (Hemagglutinin) and 9 subtypes of NA (Neuraminidase), 3 types of HA (H1, H2 and H3) and 2 types of NA (N1 and N2) have established lineages in humans [1]. The typical host of the avian influenza virus are birds and waterfowl, therefore subtypes other than those found in humans must cross the species barrier to infect humans [2].
Background
The avian influenza virus belongs to the family Orthomyxoviridae, a group of single-stranded negative sense RNA viruses with a segmented genome [3-5]. There are four major types of influenza virus: A, B, C (Gamma Influenza Virus) [5] and D (Delta Influenza Virus) [4,5]. The influenza virus primarily peaks in winter months [5]. Influenza A (IAV) infects a wide range of avian and mammalian hosts such as humans, birds, ducks, chickens, turkeys, pigs, horses and dogs. Influenza B (IBV) infects humans and seals. Influenza C (ICV) infects humans and pigs. Influenza D (IDV) was discovered in 2011 and primarily induces infection in pigs and cattle; to date no human infections have occurred [4]. Numerous studies have shown evidence that 15 hemagglutinin and nine neuraminidase subtypes of influenza are found in wild birds [6]. The IAV virus is an RNA virus with eight single stranded negative sense RNA segments. These segments encode for ten crucial proteins to carry out infection: NA, HA, PB1, PB2, PA, NP, M1, M2, NS1 and NEP/NS2 [4]. There are a few important transmembrane proteins on the viral envelope: Haemagglutinin (HA), Neuraminidase (NA) and M2 (A Transmembrane Protein). Both HA and NA are glycoproteins and are the fourth and sixth largest segments of the genome, respectively [7]. NA is an enzyme that cleaves sialic acid in order for the virus to exit the host cell after it has been infected. The NA protein is active during both the entry and exit stages. The three largest segments and the fifth largest segment out of the eight encode for viral replication machinery, which includes polymerase proteins PB2, PB1, PA (contains an endonuclease domain, which is used to seal the 5’-end cap from the nuclei of infected cells) and the nucleoprotein NP [7].,/
In some influenza A strains, the PB1 (which also contains RNA dependent RNA polymerase) segment can also encode for a second protein called PB1-F2 (an Accessory Protein) [7]. PB1, PB2 and PA together form a heterotrimeric RNA polymerase complex which is utilized during viral RNA synthesis [4]. The seventh largest segment encodes for matrix protein M1 and an ion channel protein M2 (plays a critical role in viral infection) [7]. M1 is located beneath the viral envelope and forms a matrix lattice. The M1 protein also mediates viral particle assembly [4]. The smallest segment encodes for NS1 (an interferon antagonist derived from an unspliced mRNA which functions to impair host cell antiviral responses) and a nuclear export protein [4]. The last protein required for influenza viral infection is NEP/NS2 which facilitates vRNA leaving the host nucleus to the cytoplasm. The HA protein is necessary for viral infection to occur as this protein binds to the host cell receptor and promotes membrane fusion, resulting in the virus to undergo endocytosis by the host cell [4]. In summary, endocytosis occurs first at the cell membrane, followed by the functions of the M2 protein and protons (H+) entering the cell to lower the pH. This induces the conformational change of HA which allows for membrane fusion and thus release of RNA segments (Figure1).
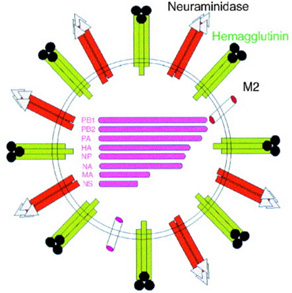
Note*: Courtesy of Suarez and Schultz-Cherry, et al. from the Immunology of avian influenza virus: a review [6].
Figure 1: Depiction of the avian influenza virus. The HA (Hemagglutinin) and NA (Neuraminidase) proteins are depicted as surface proteins in green and red, respectively. HA, NA and M2 (a transmembrane protein) can all elicit an antibody response. The 8 segments of negative sense RNA genome are depicted in the center of the virus. (MA represents the gene for the M1 protein).
The negative sense RNA segments (i.e., vRNA) are transcribed into mRNAs by the viral RNA polymerase complex. Additionally, this complex synthesizes a positive sense copy of the vRNA (cRNA) which is utilized during viral propagation in a host cell. Each vRNA segment encodes for one to three proteins [7].
Past and Present Influenza Pandemics
A pandemic is defined as an occurrence of an infectious disease worldwide or throughout a country at a particular time. During the course of the influenza virus, there have been numerous pandemics, which are outlined below. The Spanish Influenza took place from 1918-1919 and took the lives of 20-50 million people worldwide. It has now been identified that the Spanish Influenza virus was an H1N1 virus of avian origin [4,7]. The virulence of H1N1 was primarily derived from the HA protein, the replication complex, the NS1 protein and the PB1-F2 protein. However, this viral strain lacked the signature amino acids that are typically exhibited with highly pathogenic influenza strains [7]. In 1957, the Asian influenza occurred and killed an estimated of 1 million individuals worldwide. This viral strain was the result of reassortment of the human H1N1 with the avian H2N2. This reassortment resulted in a human influenza virus containing avian H2, N2 and PB1 segments [7]. The Hong Kong pandemic arose in 1968 due to the H3N2 strain [4], which had H3 HA protein and PB1 genes. Since the previous pandemic strain had the N2 protein, the death toll in the U.S. was low since individuals likely had the preexisting N2 antibody [7]. In particular, the subtypes H5 and H7 are known to cause zoonotic disease in humans with a high Case Fatality Rate (CFR) [4]. The H5N1 was first detected in Hong Kong in 1997 and has since been circulating in the local poultry and wild migratory birds of Asia, Europe and Africa [4].
Individuals from Mexico reported influenza-like symptoms and illnesses in February 2009. It was later discovered this was due to a novel H1N1 strain that was the result of a triple reassortment between human/avian/swine viruses. This H1N1 strain originated from pigs and contained six segments (PB2, PB1, PA, HA, NP and NS). Most cases of this novel strain were mild and did not require hospitalization, however risk factors such as pregnancy and obesity led to increased risk of severe respiratory disease with potentially fatal outcomes [7]. The H5N1 subtype appeared in China in 2013, causing 1,568 laboratory human infections, of which 616 were fatal [4]. While the avian influenza virus has yet to establish efficient human- to-human transmission, it is a potential pandemic threat that the virus can efficiently travel via airborne droplets [4]. Currently, two IAV subtypes and IBV subtypes co-circulate in humans (both of which are contained in the seasonal vaccine), which is the reason why there is an annual influenza vaccine.
Viral Life Cycle
There are 7 main steps of the life cycle of the influenza virus: attachment of the virus to sialic acid, endocytosis of the virus into the host cell, uncoating of the virus and membrane fusion of the virus with the host cell endosome, viral transcription, viral translation, replication of vRNA via a cRNA and lastly virion budding and release of the newly produced virion [5] (Figure2).
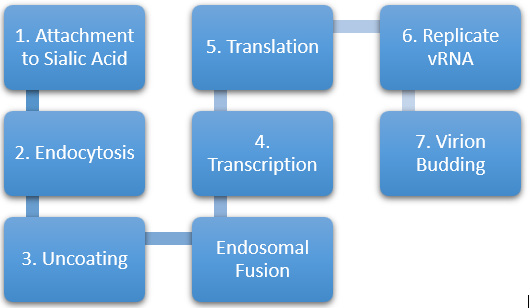
Note*: Courtesy of Hutchinson, et al. from Trends in Microbiology: Influenza Virus [5].
Figure 2: Flowchart diagram of the seven stages of the Influenza Virus life cycle.
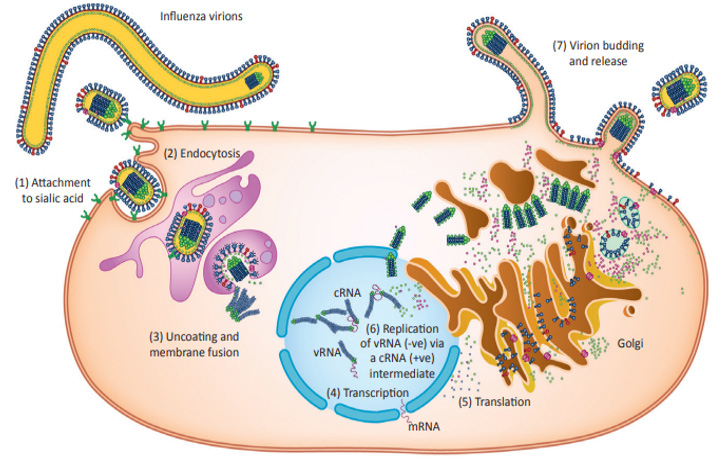
Note*: Courtesy of Hutchinson, et al. from Trends in Microbiology: Influenza Virus [5].
Figure 3: Illustration of the Influenza Virus life cycle.
The viral life cycle begins when the HA protein mediates binding of a viral particle with the host cell [4,7]. HA interacts with sialyloligosaccharides (which is present on the proteins and lipids of the host cell surface) [7]. The viral particle undergoes receptor mediated endocytosis by the host cell. The low pH in the endosome induces the HA to undergo a conformational change which allows for fusion of the viral and endosomal membranes and thus release of the viral ribonucleoprotein (vRNP) complexes (made of vRNA and NP proteins) into the cytoplasm [4,7]. The vRNP complexes enter the host nucleus, where replication and transcription lead to the amplification of the vRNA and the synthesis of mRNA to create viral proteins [5]. vRNA synthesis is mediated by the heterotrimeric polymerase complex PB1/PB2/PA [7]. This complex recognizes the promoter sequence on each vRNA segment and creates a positive sense complementary strand of each segment known as cRNA. The cRNA is used as a template strand to create more vRNA segments [4]. In the later portion of the infection cycle, newly synthesized vRNPs are exported to the cytoplasm, mediated by NEP/NS2 and M1 proteins [7]. The vRNPs are assembled with newly made viral proteins at the plasma membrane and are eventually released from the host cell as influenza virions [7]. These virions aggregate to form clusters at the cell surface because of the HA binding to sialic acids on the viral envelope and on the cell surface of the host cell [4] (Figure 3).
Influenza Virus Pathogenesis
Human influenza viruses are transmitted through the respiratory tract while avian influenza viruses are spread between bird species, primarily through direct contact in various forms such as fecal-oral, fecal-fecal and fecal-respiratory methods [4]. Both viruses predominantly replicate in the epithelial cells lining the respiratory or intestinal tract. Mild infections are associated with infection of the trachea and upper respiratory tract, whereas severe and sometimes fatal human infections occur in the lower respiratory tract. Infected epithelial cells release cytokines which initiates the inflammatory response (i.e., migration of neutrophils and macrophages to the site of infection) which thus activates more epithelial cells. This results in the release of additional cytokines such as IL-6, IL-1β, TNFα and CCL-2 which cause more epithelial damage and ultimately epithelial cell death. The avian H5N1 influenza strain causes a lethal infection in humans by initiating a hyperinflammatory response [4].
Determinants of Pathogenicity
The HA protein is a major determinant of host range of the influenza virus and its pathogenicity. The precursor protein to HA is HA0, which undergoes a posttranslational cleave to develop into two subunits: HA1 and HA2 [8]. The HA trimer (i.e., the macromolecular complex formed by three non-covalently bound proteins or nucleic acids) contains two domains: a globular head which contains the receptor binding site, and a membrane proximal stem domain. The receptor binding site is composed of three elements: 130 loop, 190 helix and the 220 loop as well as four highly conserved residues (Y98, W153, H183 and Y195). The Highly Pathogenic Avian Influenza Viruses (HPAI) contain HA proteins that possess multiple basic amino acids at the HA cleavage site. This ultimately leads to infection in poultry. In contrast, the HA proteins of avirulent influenza strains contain one basic amino acid at the cleavage site which eventually gets cleaved by a limited number of proteases that are only restricted to the respiratory or intestinal tract.
The HA protein affects which hosts become infected by the HA receptor binding specificity. Besides bat influenza viral strains (HA17 and HA18) all other HA proteins (1-16) recognize and bind to sialic acids of cell surface glycoconjugates of the host cell. The HA protein recognizes sialyloligosaccharides which are linked to galactose either via α2,6-or α2,3-linkages (indicated by NeuAcα2,6Gal or NeuAcα2,3Gal, respectively). Human influenza viruses have a high affinity for NeuAcα2,6Gal. Additionally, the human epithelial cells predominantly express the NeuAcα2,6Gal linkage and is more abundant in the upper respiratory tract rather than the lower. The Neu-Acα2,3Gal linkage is found in human lungs [8]. Human influenza viruses predominantly replicate in the upper respiratory tract, causing mild infection but efficient transmission. Avian influenza viral infection occurs more efficiently in the lower respiratory tract of humans rather than the upper respiratory tract. This results in more severe disease but less efficient transmission. Contrastingly, avian influenza viruses prefer to bind with the NeuAcα2,3Gal linkage, which is expressed in duck epithelial cells of the intestine. The epithelial cells of pigs express both NeuAcα2,3Gal and NeuAcα2,6Gal linkages. This is the reason why swine can be infected with both avian and human influenza viruses. The PB2 protein determines the pathogenicity of the influenza virus in mammalian species. Lysine at position 627 (found in human IAV and in some highly pathogenic H5N1 IAV strains) allows for efficient viral replication in the lower and upper respiratory tract of mammalian species. Glutamic acid at position 627 (found in most avian IAV strains) allows for efficient viral replication in the lower respiratory tract, but not the upper, of mammalian species.
The NS1 protein interferes with host cell antiviral immune responses by preventing the upregulation of transcription factors and interferon-β-stimulated genes. Additionally, the NS1 protein blocks the activation of viral induced RIG-I, which is a pathogen recognition receptor. The accessory protein, PB1-F2, is encoded by PB1 found in human and avian IAV viruses, but not in swine. PB1-F2 induces apoptosis by interacting with the mitochondrial proteins and by interacting with the PB1 segment to retain viral ribonucleoprotein complexes in the nucleus of the host cell. One of the reasons that contributed to the high virulence of the H5N1 virus of the Spanish Influenza in 1918 is probably a single amino acid change in the PB1-F2 protein, demonstrating the effect of PB1-F2 on the virulence of influenza virus.
Mechanisms of Influenza Virus Evolution
The mechanism of evolution of the influenza virus is mediated by two methods: antigenic drift and antigenic shift [4]. Antigenic drift occurs with the accumulation of multiple point mutations in the viral genome during replication. These mutations arise by a lack of proof reading from the viral RNA-dependent RNA polymerase. Mutations in the HA and NA proteins lead to decreased recognition of pre-existing antibodies that were used against previous viral strains. Mutations in HA and NA proteins has led to the prevalence of multiple variants. Antigenic drift is the reason for the occurrence of seasonal influenza booster vaccines [4]. From recent studies, it appears that chickens are not as sensitive to antigenic drift as humans, however the mechanism behind this finding is unknown [6]. One possible explanation highlights the host and host immune response likely play a large role in this novel finding [6]. Antigenic shift is the result of reassortment between viral RNA segments of two different influenza viruses co-infecting the same host cell [4,8,9]. This reassortment leads to the development of new strains which can elicit severe infections or spread quickly through a population that does not display pre-existing immunity. In fact, the 1957 Asian Influenza Pandemic was the result of reassortment between H2N2 avian virus and H1N1 human virus. The 1968 Hong Kong Influenza Pandemic resulted from the reassortment of H3 avian influenza and H2N2 human influenza virus [4].
Clinical Presentations
Individuals infected with H5N1 or H7N9 (i.e., the avian influenza strains) can experience mild to severe symptoms. These symptoms are similar to those associated with the seasonal flu, such as cough, fever, sore throat, nasal congestion, muscle or body aches, fatigue, headaches, shortness of breath and conjunctivitis [10]. Type A and B influenza viruses cause respiratory infections that either manifest as asymptomatic in young or healthy individuals, and may be life-threatening in some, especially older patients and those with comorbid or immune suppressed conditions. Symptoms include cough, fever, sore throat, headache, chills, anorexia, malaise, coryza and myalgia. The incubation period ranges from 1-4 days followed by a symptomatic period of 3-4 days. Individual outcome of the infection is dependent on multiple factors, such as pre-existing immunity, biological properties of the virus and the health status of the individual prior to infection. Certain risk factors such as heart or lung disease, immunological disorders, renal failure and smoking increase the risk of the infection becoming more severe [7]. The H3N2 influenza virus poses the most severity among the influenza viruses, then influenza B viruses and lastly the H1N1 virus. Influenza C viruses typically do not cause disease symptoms [7].
Immunology: Host Immune Responses
A fine-tuned immune response can eliminate the viral infection, however a prolonged inflammatory response results in tissue damage and a severe viral infection. Upon entry into the host, the influenza virus is recognized by the innate immune system through a myriad of pattern recognition receptors (PRRs), toll-like receptors (TLR3, TLR7 and TLR8), cytosolic receptor RIG-I and NOD-like receptor NLRP3. Cytokines and upregulation of IFN-I/III (interferons I and III) occurs after the activation of TLR and RIG-I signaling pathways. IFN-I/III act via paracrine and autocrine signaling to induce expression of the Interferon Stimulating Genes (ISGs) which exert antiviral activity to restrict viral replication. Cytokines also activate phagocytes which recruits more immune cells to the site of infection, ultimately contributing to adaptive immunity. NLRP3 inflammasome (a multiprotein complex which functions to regulate the innate immune response and inflammatory signaling) activates the secretion of cytokines IL-1β and IL-I8 which induces pyroptosis (inflammatory cell death caused by microbial infection) of infected cells [4] (Figure 4).

Note*: Courtesy of Liang Yuying, et al. Pathogenicity and Virulence of Influenza [4].
Figure 4: The following RTK receptors are utilized by the IAV virus for replication: TrkA, c-Met and EGFR, all of which are located at the host cell membrane.
Macrophages play an important role in the innate immune response by secreting cytokines [6,4] to initiate the adaptive immune response and clearing debris and dead cells [4]. Additionally, macrophages function by utilizing phagocytosis to engulf foreign particles [6]. When macrophages utilize phagocytosis, they produce oxygen free radicals and Reactive Oxygen Species (ROS) [6]. ROS metabolites result in tissue damage at the site of inflammation [6]. This contributes an explanation for why animals infected with the influenza virus experience secondary bacterial infection [6]. Alveolar macrophages reside in tissue. However, an excessive amount of cytokine secretion from macrophages can lead to lung damage and worsening of the disease severity. This highlights the importance of regulating macrophage activation as it is critical for protection against influenza viral infections. Previous studies have shown cellular Wnt/β-catenin/HIF via the -1α signaling pathway and transcriptional factor PPAR-γ promote inflammatory activity of alveolar macrophages. Downregulation of this transcription factor allows for the suppression of an excessive inflammatory response [4].
There are two main classes of macrophages: proinflammatory M1 and anti-inflammatory M2. M1 macrophages are activated early in the infection cycle to secrete inflammatory cytokines, such as IL- 1, IL-6 and TNF-α, all of which trigger an antiviral response. M2 macrophages are utilized in the later portion of the infection cycle to terminate inflammation, repair tissue damage and produce TGF-β (transforming growth factor-beta is a multifunctional cytokine) and IL-10. Recent studies have shown that M2 macrophages show more benefit in those infected with the influenza virus versus the M1 macrophages, suggesting a solution for a future novel therapy against the virus [4]. Macrophages are highly susceptible to infection by the influenza virus. The highly pathogenic H5N1 virus is known to infect alveolar macrophages [4].
Two types of T cells, CD4 and CD8, are components of the adaptive immune response and function to clear viral infections and prevent the severity from worsening of diseases. Pre-existing influenza specific CD4 T cells are commonly associated with lower viral shedding (i.e., lower rate of viral replication) and less severe disease in health individuals following the infection. CD8 T cells play a crucial role in memory against the influenza virus. Patients with early recovery from the H7N9 viral infection displayed H7N9-specific CD8 T cells; those with prolonged hospitalization of the same virus displayed late recruitment of CD4 and CD8 T cells. This suggests that an early strong memory T cell response prevents or reduces disease severity. However, overactivation of T cells can lead to tissue damage due to hyperinflammation [4]. Tissue-Resident Memory T cells (TRM) are part of the first line of defense against an infection. Lung CD8 TRM facilitates protection against respiratory infection by producing effector responses at the site of infection immediately upon infection. However, heightened responses lead to inflammation and tissue damage [4].
Host Cell Signaling Factors and Pathways Contribute to Viral Replication
All viruses, including the influenza virus, utilize host cell machinery and host factors to replicate. There are host cell factors that promote the endocytosis of the virus, such as Free Fatty Acid Receptor 2 (FFAR2) and transmembrane protein Immune Globulin Superfamily DCC subclass member 4 (IGDCC4). Once endocytosis is complete, the viral particle moves from the early to late endosome where fusion of the virus to the endosomal membrane takes place as well as the release of vRNPs. The Cysteine Protease Cathepsin W (CtsW) and lysosomal Acid Phosphatase 2 (ACP2) from the host cell are necessary for fusion of the virus with the endosome to occur. It is known that the epidermal growth factor receptor pathway substrate 8 (EPS8) promotes influenza viral uncoating [4]. After endocytosis, the RNA segments must travel from the cytoplasm to the nucleus of the host cell so viral RNA synthesis and replication can occur. Afterwards, viral protein synthesis and particle assembly occurs in the cytoplasm. The influenza virus utilizes the cellular nucleocytoplasmic trafficking machinery for nuclear import into the host nucleus as well as for export of vRNPs, viral mRNAs and viral proteins. Gaining access to the host nucleus is enhanced by G protein subunit β1 (GNB1), which functions to increase the interaction between PB2 and importin proteins. BinCARD1 increases the binding of NP with importin α7, which also facilitates entry of the RNA segments into the nucleus. Additionally, the 5’-end cap is sealed from cellular mRNA. Nuclear export of viral mRNA uses the host mRNA export machinery to translocate the viral mRNA via the nuclear pores to the cytoplasm [4].
There are also host factors that interact with viral matrix proteins M1 and M2. M1-interacting host factors, such as GNB1, bind to M1 on the plasma membrane and promote viral release [4]. The influenza virus, in addition to interacting with viral host factors, also utilizes host cellular pathways which are subsequently used for viral replication. Receptor Tyrosine Kinases (RTKs) and their downstream signaling pathways (Raf/MEK/ERK, Phosphatidylinositol- 3-kinase (PI3K)/Akt, JAK/STAT, PLC-γ1, NF-kB) facilitate viral replication. The influenza virus activates many RTKs, such as epidermal growth factor receptor (EGFR), c-Met and Tropomyosin Receptor Kinase A (TrkA). It is known that once EFGR and c-Met are activated, they both promote viral uptake quickly. Additionally, activation of EGFR suppresses IFN-I/III production and antiviral responses, which further increases viral production [4]. TrkA has a high affinity for its predisposed ligand, Nerve Growth Factor (NGF) which plays a role in neuronal cell development and function. TrkA is typically expressed in non-neuronal tissues, one of which includes the human lung epithelial and endothelial cells, both of which play a role in airway inflammation. TrkA promotes viral replication in the airway epithelial cells and contributes to airway inflammation by activating the expression of proinflammatory cytokines in infected cells [4].
Transmission of the Avian Influenza Virus to Humans
Human influenza viral infections typically occur because of direct contact between viruses from infected birds to humans [3]. It is known that IAV virus subtypes H1N1, H3N8, H3N2, H6N2, H6N1, H9N2, H4N8 and H10N7 have the ability to replicate in humans [8]. This replication was experimentally tested in 81 healthy individuals. Those infected with H4N8, H10N7 or H6N1 experienced mild viral replication in the nasopharynx with mild upper respiratory symptoms. However, attempts to transfer H6N1 from one infected individual to a healthy participant were not successful. Those infected with H1N1, H3N2, H3N8, H6N2 or H9N2 had no evidence of viral replication in the nasopharynx nor symptoms. These results suggest that subtypes H1 or H3 and N1 or N2 provided cross-reacting immunity to prevent avian viral replication [8]. Besides the H5N1, there are other avian viruses that cause disease in humans, such as H7N7, H9N2 and H7N3. These subtypes are largely known to also cause disease in terrestrial poultry. Terrestrial poultry, such as chickens or ducks, possess the NeuAcα2,6Gal receptor which bind to human influenza viruses. This suggests that the H9N2 avian viral subtype can bind to multiple hosts including humans and terrestrial poultry species [8].
The first human infection caused by H5N1 was in 1997 at Hong Kong, which resulted in 18 cases and six deaths. The source of these infections was live-poultry markets where ducks, geese, chickens and other species that were sold for later human consumption. In February 2003, a father and son became infected with H5N1 which resulted in severe acute respiratory disease from the People’s Republic of China. Evidence indicated that there was no mutation in the stalk of the NA protein. However, there was an amino acid replacement at position 227. This mutation allowed the virus to bind to both the avian SA-α2,3 receptors and the human SA-α2,6 receptors, which did not change the ability of the virus to elicit human-to-human transmission [8]. Most human cases of H5N1 resulted from direct contact or handling with infected poultry, slaughtering infected poultry or preparing them for consumption. Other cases resulted from contact with infected raw blood, even when the poultry appeared to be asymptomatic. The portal of entry between bird-tohuman transmission is likely the respiratory tract, gastrointestinal tract or the conjunctiva (mucous membrane that covers the front of the eye) [8]. Wild birds are suspected of being a vector of the H5N1 avian virus. Particularly, wild waterbirds are suspected since the virus spreads through areas that had no record of the virus’ presence and this coincided with the migration of the wild waterbirds between those areas [11]. In 2006, H5N1 was detected in multiple wild waterbirds in western Europe, primarily in areas that had no record of the virus. The main argument against wild waterbirds being the vector is that upon their infection, the animal would quickly die from infection or was too sick to travel a long distance to transmit the disease to humans [11].
Transmission Within Poultry and Wild Birds
Live-poultry markets found in Asia, especially where unsold live poultry are held overnight in the market, allow the avian influenza virus to maintain replicating and thus amplify within the poultry. Virus isolation rates in this poultry is higher than those detected in incoming poultry. Virus transmission can be interrupted with a “rest day” in which the market is completely eradicated of poultry. While the virus can replicate rapidly in live-poultry markets, they can also undergo effective transmission via empty cages to other poultry farms [8]. In April 2023, the CDC discovered two novel subtypes of the avian H5N1 virus of clade 2.3.4.4b HA. It was determined that the new subtypes were 99% identical to the original, highly pathogenic H5N1 subtype, suggesting no evidence of genetic reassortment via antigenic shift. These novel subtypes were discovered in the majority of wild birds in South America [12].
Atypical Avian Influenza Case
The CDC reported the first case on March 9, 2004, of avian influenza in Ayudhaya in central Thailand. The patient was a 39-year- old female who initially displayed no respiratory symptoms but had a fever and diarrhea [1]. It is known that unlike human influenza viruses that predominantly replicate in the respiratory tract, avian influenza viruses can replicate in the gut epithelial cells [13]. Additionally, the patient had no history of pre-existing diseases. She was transferred to the hospital due to rapidly progressing pneumonia and a progressive fever for 1 week, along with diarrhea, nausea and vomiting with no early respiratory symptoms [1]. Initial lab results included: leukocyte count 3300 cells/mm3 (4,500- 10,500 cells/mm3 is normal), total lymphocyte count 640 cells/ mm3 (1,000-4,000 cells/mm3 is normal), hemoglobin 13g/dL (12- 16g/dL is normal) and platelet count 400,000cells/mm3 (250,000- 260,000 cells/mm3 is normal). Because the patient was presented with no initial respiratory symptoms, a chest radiograph was not performed. Norfloxacin was prescribed. On day 5 in the hospital, the patient developed a cough and shortness of breath and thus a chest radiograph was performed, which indicated patchy infiltration at bilateral lower lung regions [1]. Viral pneumonias typically display bilateral patchy infiltrates on X-Rays [8].
The patient was transferred to a second hospital, upon which her temperature was 39.4 C (102.92F), respiratory rate 44breaths/ min, blood pressure 110/80mmHg and heart rate 140beats/minute. The following day, the infectious disease department was asked to consult [1]. The patient’s family revealed that she was exposed to several dead chickens in her neighborhood. She was tested for H5N1 influenza via the rapid influenza A test by enzyme immune membrane filter assay, which yielded a negative result. Thus, NA inhibitors were not prescribed. The following day, the patient died due to multiorgan failure and severe adult respiratory distress syndrome. Nasopharyngeal aspirates were positive for influenza A H5 strain. Additionally, several dead chickens in the patient’s neighborhood tested positive for H5N1 via viral cultures [1].
Summary
The avian influenza virus comes in various subtypes, the most prominent of which is H5N1 that causes human viral infections. Most human infections occur due to contact with infected poultry, although the mechanism of human-to-human transmission is not clearly understood. The virus has many bird and mammalian hosts, such as wild ducks, shorebirds and gulls. Once a human is infected, the virus can cause fatal symptoms as part of HPAI or MPAI. The four main classes of influenza are A, B, C and D, each of which infects different host species. The glycoproteins that give rise to various subtypes of this virus are NA and HA.
Acknowledgements
None.
Conflicts of Interests
None.
References
- A Apisarnthanarak, R Kitphati, K Thongphubeth, P Patoomanunt, P Anthanont, et. Al. (2004) Atypical Avian Influenza (H5N1). Emerg Infect Dis 10(7): 1321-1324.
- Nicholson KG, Wood JM, Zambon M (2003) Influenza. Lancet 362(9397): 1733-1745.
- Samson SY Wong, Kwok yung Yuen MD (2006) Avian Influenza Virus Infections in Humans. Chest 129(1): 156-168.
- Liang Yuying (2023) Pathogenicity and virulence of influenza. Virulence 14(1): 2223057.
- Edward C Hutchinson (2018) Influenza Virus. Trends Microbiol 26(9): 809-810.
- DL Suarez, S Schultz Cherry (2000) Immunology of avian influenza virus: a review. Dev Comp Immunol 24(2-3): 269-283.
- Kawaoka Yoshihiro, Neumann Gabriele (2012) Influenza Viruses: An Introduction. Methods Mol Biol 865: 1-9.
- JS Malik Peiris, Menno D de Jong, Yi Guan Avian (2007) Influenza Virus (H5N1): a Threat to Human Health. Clin Microbiol Rev 20(2): 243-267.
- John Biegel, Mike Bray (2008) Current and Future Antiviral Therapy of Severe Seasonal and Avian Influenza. Antiviral Res 78(1): 91-102.
- NIH National Library of Medicine (2017) Bird Flu. Medline Plus.
- Keawcharoen Juthatip, Van Riel Debby, Van Amerongen Geert, Bestebroer Theo, E Beyer Walter, et. al. (2008) Wild Ducks as Long-Distance Vectors of Highly Pathogenic Avian Influenza Virus (H5N1). Emerg Infect Dis 14(4): 600-607.
- (2023) Human Infection with Highly Pathogenic Avian Influenza A(H5N1) virus in Chile. Centers for Disease Control.
- Xuejiao Han, Luca D Bertzbach, Michael Veit (2019) Mimicking the passage of avian influenza viruses through the gastrointestinal tract of chickens. Vet Microbiol 239: 108462.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.