Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
RNA-Seq Workflow Optimization for Heavily Degraded FFPE Samples with GLP & GCLP Compliance
*Corresponding author: Yixiao Cui, Frontage Laboratories, Inc., 760 Pennsylvania Drive, Exton, PA, USA
Received: May 03, 2024; Published: May 14, 2024
DOI: 10.34297/AJBSR.2024.22.002977
Abstract
Regulated studies with formalin-fixed paraffin-embedded (FFPE) samples for Investigational New Drug (IND) application or New Drug Application (NDA) submission, have been emerging to tightly follow GLP (good laboratory practice) and GCLP (good clinical laboratory practice) standard, especially for next-generation sequencing (NGS) readouts that are indispensable for genetic medicine/gene therapy. However, in addition to the sophisticated workflow of NGS, RNA degradation (DV200<20%) in FFPE samples, is notoriously challenging. Here, we report a standardized workflow for RNA-Seq of FFPE samples with rigorous Good Document Practice (GDP), particularly, underscoring the optimization procedure of the sequencing library construction. We obtained 4 normal human tissue FFPE samples with DV200 values ranging from 8.15% to 15.44%, including human liver, tonsil, thymus, and pancreas tissues. Intriguingly, applied our library construction procedure, the ratio of “good libraries” has been significantly increased by 25%-65%. Of note, our integrated workflow, featured with increasing RNA input concentrations, decreasing the hybridization temperature from 94°C to 70°C for rRNA depletion, skipping the fragmentation and denaturation step, and adding additional PCR, enables us constructing high-quality libraries, characterized by around one to two times total reads increasement and 1.3 to 6.5 times exon reads yield. Thus, we accomplished an optimized RNA-Seq library preparation for degraded FFPE samples. Taken together, our RNA-seq workflow for degraded FFPE samples with GLP and GCLP compliance holds great promise for a broad application potential in regulated preclinical and clinical studies.
Keywords: FFPE, NGS, RNA, GLP, Genomics, Library construction
Introduction
Formalin-fixed, paraffin-embedded (FFPE) tumor tissue is the most widely used method of preserving nucleic, protein and histology for diagnostics and research purposes [1]. FFPE samples have many advantages for application in genomics research, like FFPE sections displaying various historical features of cancer, including precancerous lesions, enable assessment of the genetic events related to the observed histological change, FFPE tissue samples allow for a retrospective study, with increases in the number of cancer case and types [2,3]. In addition, FFPE is the ubiquitous room temperature clinical tissue biospecimen preservation method. However, there are several types of DNA and RNA damage in for malin-fixed tissues [4], including 1) DNA fragmentations; 2) formaldehyde- induced crosslinks; 3) generation of a basic site; 4) deamination of cytosine bases leading to C->T mutations; and 5) RNA degradation. This will present notable challenges when purifying DNA or RNA from FFPE samples due to biomolecule crosslinking, nucleic acid fragmentation, degradation, and low yield [5,6]. These difficulties also impose significant demands on the analysis techniques used for subsequent samples.
Next-generation sequencing (NGS) offers tremendous discovery capabilities for detecting novel or rare variants and generating high-throughput data [7]. This powerful tool is evolving rapidly and plays an important role in drug discovery and development [8], cancer diagnostics, pathogen identification, and precision medicine. NGS has a wide range of applications, including whole-genome sequencing (WGS) [9,10], whole-exome sequencing (WES) [11], whole transcriptome shotgun sequencing (WTSS)-also known as RNA sequencing (RNA-seq) [12,13], targeted/candidate gene sequencing (TS) [14], and methylation sequencing (MeS) [15]. To ensure good data yield, there are challenges encountered during NGS operations, especially with poor quality RNA or DNA material, which is a common scenario for clinical samples. Previously, there were hardly any studies or methods to guide how to maximize the acquisition of effective genetic information for sequencing analysis in situations of extremely low RNA quality, especially concerning poor-quality FFPE slides samples.
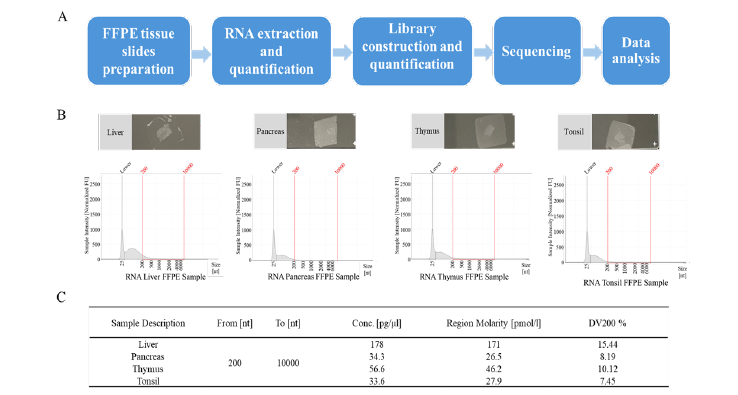
In this study, we established a standardized RNA extraction method from FFPE slides and an RNA-seq operation method adhering to Good Lab Practice (GLP) standards (Figure 1A) [16,17], which have been consistently able to get high-quality data, particularly for degraded RNA (DV200<20%) from FFPE. We also presented some key skills, including the quantification, library kit selection, library construction, additional post-PCR process and data analysis. Our research will empower the use of FFPE samples in NGS without the need to account for materials quality, thus expanding the application of NGS across diverse fields.
Methods
FFPE Tissue Slides
We obtained 4 normal human tissue samples, comprising human liver, tonsil, thymus, and pancreas tissues, from BioIVT. All samples underwent routine fixation in 10% neutral buffered formalin and embedding in paraffin. The FFPE blocks were stored at ambient/room temperature. Sections measuring 3-5 μm thin were cut from the FFPE blocks and placed on positively charged glass slides. These slides have been maintained at ambient/room temperature for over 6 months since cutting.
RNA Extraction
RNA was extracted from FFPE tissue slides samples using the RecoverAllTM Multi-Sample RNA/DNA Workflow (ThermoFisher Scientific) according to the manufacturer’s protocol. All RNA samples were dissolved in 30 μL pre-heated elution buffer and stored at -80°C.
Library Construction
Sample libraries were constructed using Illumina Stranded Total RNA Prep, Ligation with Ribo-Zero Plus (Illumina) according to the reference guide. The RNA input, fragmentation, denaturation and PCR steps are optimized, and described in the results section. IDT for Illumina DNA/RNA ID Index set A and IDT for Illumina RNA index Anchors were used for the library construction.
Quality Assessment of RNA and Library
The concentration of RNA and libraries were calculated by QubitTM Flex Fluorometer (ThermoFisher Scientific). The measurement of the RNA and libraries fragment sizes were done using 4200 TapeStation system (Agilent Technologies).
Next Generation Sequencing
NextSeq550Dx instrument was used for the sequencing. The NextSeq 500/550 High Output (300 Cycles, up to 400 million reads) reagent cartridge (Illumina) was used for the experiment, and pairend sequencing was applied for the assay. 1% PhiX control was added to the sequencing.
Data Analysis
Trimmomatic (Version 0.39) tool was used for removing adapter sequences, primers, and other types of unwanted sequences from the high-throughput sequencing reads. Sequence quality scores, base content, sequence duplication levels, adapter contamination were obtained using Fast QC (Version 0.11.9) tool. HISAT2 (Version 2.2.1) tool was used for aligning RNA -seq reads to the reference genome. Feature Counts (Version 2.0.3) tool was applied to RNA sequencing data to count the number of reads that align with the reference genome.
Results
RNA Quality of FFPE Tissue Slide Samples
We extracted RNA from tonsil, thymus, spleen, pancreas, and liver tissue FFPE slides (Figure 1B) and determined RNA quality using TapeStation. These FFPE slides samples are 3μm thin and have been fixed in 10% NBF formalin, processed, and then paraffin embedded into blocks. They were all stored at ambient/room temperature for more than 6 months before processing. Due to long-term storage at room temperature, some RNA will be heavily degraded. The percentage of RNA fragments longer than 200 nucleotides (DV200%) is a parameter used to assess the quality of RNA samples, 70% or higher is considered indicative of high-quality RNA and a DV200 value below 70% is often considered indicative of RNA degradation and lower RNA quality. In addition, the RNA input recommendations in library construction suggest that FFPE samples’ DV should be higher than 55%. These RNA samples’ DV200 values are very low, ranging from 8% to 16% (Figure 1C). There is almost no presence of RNA peaks in the region greater than 200bp (Figure 1B). Due to the low quality of the RNA, this will pose significant challenges for the subsequent library construction.
Therefore, optimizing and improving the conditions to enhance the quality of the library are particularly important.
Optimization of Library Construction
The quality of the library is crucial to sequencing, as its condition directly affects the quality of data and the analysis results of the experiment. Although the low quality of RNA due to the sample storage or preparation may occur, optimizing the conditions for the library construction can significantly improve the quality of sample data and yield more valuable genetic information. The process of library construction is very tedious. There are several key steps in this process that can affect the quality of the library, and they are our focus for attention and optimization (Figure 2A). First, we improve the RNA input concentrations, and more RNA will lead to more data information. Second, for the rRNA depleting step, we decrease the hybridization temperature from 94°C to 70°C. High temperature will damage the RNA structures, causing them to be fragmented and degraded. However, if the temperature is too low, the RNA cannot open their own structures and combine to the probes, hindering the removal of rRNA. Then, for the fragmentation and denaturation of RNA step, we skip the “94°C for 2 mins” step. Most of our RNA is below 200 bp according to the TapeStation results (Figure 1B) and we don’t need to denature and fragment the RNA again. In addition, library PCR cycle number is another key factor during the library construction, too few PCR cycles will result in a low library yield, incomplete amplification, and loss of genetic information, whereas too many PCR cycles can lead to an increase in amplification error rate, resulting in inaccurate genetic information. According to our RNA sample quality and concentration, we increased the number of PCR cycles by two more than usual during the experiment.
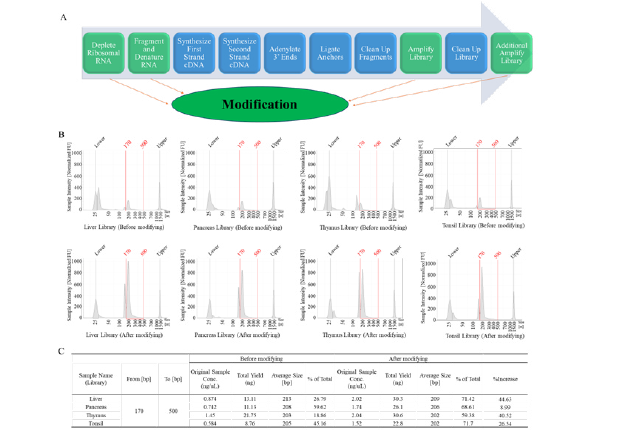
Figure 2: The flow of library construction and the libraries of FFPE samples. A, the entire process of library construction and modification. B, the libraries quality testing results using TapeStation. C, the quality and yield of the library before and after condition optimization.
We compared the quality and yield of the library before and after condition optimization (Figure 2B and Figure 2C). Before the optimization, the yields of the liver, pancreas, thymus, and tonsil sample libraries ranged from 8.76 ng to 21.75 ng and the ratio of good library with fragment sizes between 170 bp to 500 bp is 18.86% to 59.62%. After the optimization, the yields of libraries have been increased by 1.5-3 times in comparison with routine workflow and the ratios of the good library have been increased by 9% to 45%. For the low-quality libraries, such as the liver and thymus libraries, the ratios of the good libraries can be increased by more than 40% after method optimization.
Optimization of additional PCR conditions
After optimizing the library construction process, we found that the proportion of “good library” did not reach the expected threshold of over 80%. This will affect the quality of sequencing, resulting in a high proportion of invalid data. To improve the “good library” ratios, we used P5, P7 primers to the second-round library PCR and optimized the PCR conditions.
Annealing temperature: The annealing temperature is a critical factor during PCR. If the temperature is too high, primers cannot bind efficiently to the target sequence, resulting in low amplification. Conversely, if the temperature is too low, there is a higher likelihood of nonspecific amplification occurring. According to the primers’ Tm, we tested 60°C, 65°C and 70°C annealing temperature. As the temperature rises, the proportion of fragments between 170 bp and 500 bp will initially increase and then decrease, reaching its highest point when the annealing temperature reaches 65 degrees (Figure 3A and Figure 3B). So, we decided to set the annealing temperature to 65°C first.
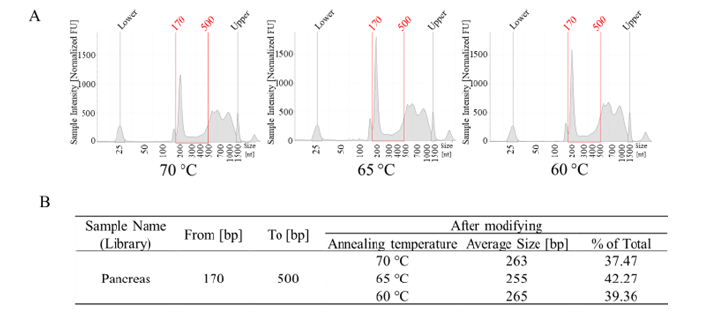
Figure 3: PCR annealing temperature optimization results. A, the libraries quality testing results using Tape Station. B, the library average size and “good library” ratio of different annealing temperature.
PCR cycle numbers: Properly increasing the number of PCR cycles can enhance the sample yield; however, excessive PCR cycle numbers can also generate many non-specific products. For our previously optimized process, the proportion of good libraries has exceeded 50% and further amplification at a certain cycle will lead on a further increase in the proportion of good library. This achieves the effect that cannot be attained solely by increasing the PCR cycles during library construction. It underscores the necessity of performing secondary PCR of the products and optimizing conditions. We have tested PCR cycles ranging from 6 to 10 and found that the proportion of “good library” also exhibited a trend of initially increasing and then decreasing (Figure 4A and Figure 4B). When the PCR cycle number was 7, the proportion of the pancreas sample library between 170 bp and 500 bp reached 87.16%. After the initial optimization (Figure 2C), the proportion of “good library” increased by 8.99% followed by an additional increase of 18.55%, resulting in a total increase from 59.63% to 87.16% (Figure 4B).
All sample applications: We did secondary optimization on the library products of all samples, the proportion of “good library” for all samples can reach 80% or more (Figure 5A and Figure 5B). According to the results, even if some samples have very poor quality with a low proportion of high-quality libraries, after two rounds of optimization experiments, the improvement in library quality can exceed 60%. For example, in the case of the thymus sample, the proportion of “good library” before optimization was only 18.96%, but after optimization, it reached 82.64%. This will significantly improve the quality of sequencing.
Data Analysis
We upload samples with the same concentrations, yet the total reads of different samples vary (Figure 6C). Due to the poor quality of the libraries, the Cluster Passing Filter rate is only 66.02%. In normal circumstances, the Cluster Passing Filter rate should be above 75%. We performed simple statistical analysis on the mapped reads and exon reads counts using our pipeline (Figure 6A). Compared to the number of total reads before the improvement of the library, it has increased by about 1 to 2 times after the method improvement (Figure 6B and Figure 6C). The number of exon reads has increased by 1.3 to 6.5 times compared to before method optimization and the proportion of the exon reads number also increased (Figure 6B and Figure 6C).

Figure 4: PCR cycle number optimization results. A, the libraries quality testing results using TapeStation. B, the library average size and “good library” ratio of different PCR cycle number.
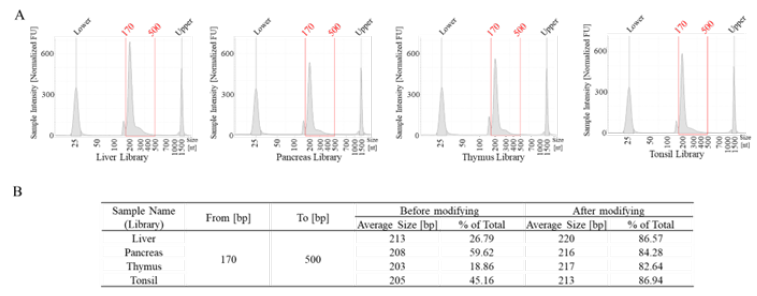
Figure 5: All sample optimization results. A, the libraries quality testing results using TapeStation. B, the library average size and “good library” ratio after method optimization.
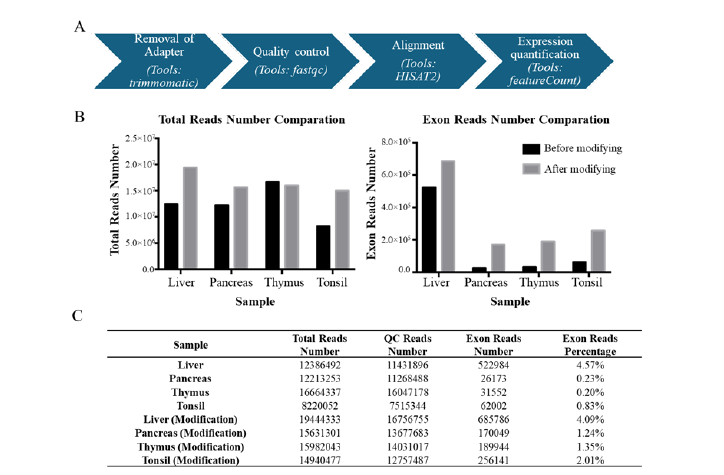
Figure 6: The RNA-seq data analysis pipeline and results. A, the exon reads number analysis pipeline. B, the total reads number comparation before and after modification (left) and the exon reads number comparation before and after modification (right). C, the total, QC and exon reads count statistics.
Discussion
FFPE slides samples are commonly used in histology and pathology for various diagnostic and research purposes in drug discovery and development [18]. FFPE slides preparation only requires a very small amount of tissue samples, and they can be stored at room temperature for extended periods. For many low-volume and extremely precious samples FFPE slides are the preferred method of preservation. However, due to the utilization of various organic reagents, wax, and other substances during the preparation process, there will inevitably be some impact on the quality of the samples. Since it is not possible to improve the quality of the samples themselves, enhancing and improving the pre-sequencing processing techniques of the sample is necessary to obtain more accurate genetic information. The quality of sequencing generally depends on the following factors: the quality of the DNA/RNA; the quality of the library; the standardization of the on-machine operations.
It poses significant challenges for extracting DNA/RNA and obtaining effective information subsequently. In the experiments, our samples have been stored at room temperature for at least 6 months, and some RNA is degraded heavily with the DV% of only 8.15% to 15.44% (Figure 1B and Figure 1C). Most commercial library construction kits can only be used with “good quality RNA” which DV% should be over 55%. Following our studies on the mechanism of library construction and the characteristics of RNA, we found that “temperature” has a significant impact on the quality of library. Therefore, we cleverly lowered the temperature, skipping or adding certain steps to increase the proportion of “good library”. For the improvement, we skipped “94°C for 2 mins” in the RNA fragmentation and denaturation step. The average length of the library has been increased without subjecting it to “94°C for 2 mins” (Figure 5B).
Our studies suggest that not all library constructions require denaturation treatment; rather it really depends on the quality of the initial sample to determine whether fragmentation is necessary. In cells, the abundance of ribosomal RNA (rRNA) typically ranges from 80% to 90%. However, we need to remove the rRNA during library process because they do not carry genetic information. According to the quality and the structure of the RNA, we decrease the hybridization temperature from 94°C to 70°C. Based on the number and proportion of exons we observed, we confirm that lowering the temperature did not affect the removal of rRNA, 70°C is sufficient to open the structures of RNA and probes, allowing them to bind together and achieve the removal of rRNA. Together, these steps enable us to effectively remove the interference of rRNA and generate a high-quality RNA library, allowing us to obtain more genetic information.
PCR is an essential process in constructing libraries and there are two key factors affecting the whole processer: annealing temperature and the number of PCR cycles. A too high annealing temperature may lead to decreased amplification efficiency, while a too low annealing temperature may result in non-specific amplification (Figure 3A and Figure 3B). Also, a low number of PCR cycles can result in a low yield of the library, which may not be sufficient for subsequent steps (Figure 4A and Figure 4B). However, if the number of PCR cycles is too high, it can lead to significant non-specific amplification or an increase in mismatch rates, thereby reducing the accuracy of sequencing information. According to the principle of PCR amplification, the copy number will increase exponentially. This implies that if the proportion of “good libraries” can exceed 50% in the first round of amplification (Figure 2C), the proportion of “good libraries” will further increase substantially in the subsequent second round of amplification. In this study, we utilized two rounds of amplification through conditional optimization, resulting in elevating the proportion of “good library” to over 80% (Figure 5B). Additionally, we conducted further validation of the method’s effectiveness using sequencing data. Exon analysis demonstrated that after implementing the optimization method, there was a significant increase in both the number and proportion of obtained exons (Figure 6A and Figure 6B). Hence, our optimization protocol has demonstrated significant benefits in extracting valuable information from low-quality samples.
Conclusion
We effectively optimized and bolstered the sequencing quality of FFPE samples through strategies such as augmenting RNA input, refining the library construction process, and incorporating supplementary PCR steps. This led to a substantial enhancement, with effective information increasing by 25% to 61%. Moreover, we instituted rigorous GLP standards for conducting RNA-seq experiments with well documented assay procedures. Our findings hold promise to streamline the utilization of FFPE sample materials in NGS, alleviating concerns regarding material quality and consequently broaden the scope of NGS applications across diverse fields.
References
- Lin Y, Dong Z H, Ye TY, Luo JC, Gao J, Guo AY, et al. (2024) Optimization of FFPE preparation and identification of gene attributes associated with RNA degradation. NAR Genomics and Bioinformatics 6(1): lqae008.
- Ahlfen S, Missel A, Bendrat K, Schlumpberger M (2007) Determinants of RNA Quality from FFPE Samples. PLoS ONE 2(12): e1261.
- Zvenigorosky V, Gonzalez A, Veith G, Christine Keyser, Christian Bonah, et al. (2024) Evaluation of whole-genome enrichment and sequencing of T. pallidum from FFPE samples after 75 years. Iscience 27(1): 108651.
- Belder N, Coskun Ö, Erdogan BD, et al. (2016) From RNA isolation to microarray analysis: comparison of methods in FFPE tissues. Pathology-Research and Practice 212(8): 678-685.
- Einaga N, Yoshida A, Noda H, et al. (2017) Assessment of the quality of DNA from various formalin-fixed paraffin-embedded (FFPE) tissues and the use of this DNA for next-generation sequencing (NGS) with no artifactual mutation. PLoS ONE 12(5): e0176280.
- Arreaza G, Qiu P, Pang L, et al. (2016) Pre-analytical considerations for successful next-generation sequencing (NGS): challenges and opportunities for formalin-fixed and paraffin-embedded tumor tissue (FFPE) samples. International journal of molecular sciences 17(9): 1579.
- Kwon R & Yeung CC (2024) Advances in next-generation sequencing and emerging technologies for hematologic malignancies. Haematologica 109(2): 379-387.
- Abdi G, Tarighat MA, Jain M, et al. (2024) Revolutionizing Genomics: Exploring the Potential of Next-Generation Sequencing. In Advances in Bioinformatics. Singapore: Springer Nature Singapore pp: 1-33.
- Mustafa AS (2024) Whole Genome Sequencing: Applications in Clinical Bacteriology.
- Bagger FO, Borgwardt L, Jespersen AS, et al. (2024) Whole genome sequencing in clinical practice. BMC Medical Genomics 17(1): 39.
- Khan WA (2024) Whole-exome and whole-genome sequencing in the molecular diagnostic laboratory. In Diagnostic Molecular Pathology. Academic Press pp: 27-38.
- Dufresne A, Attignon V, Ferrari A, et al. (2024) Added value of whole‐exome and RNA sequencing in advanced and refractory cancer patients with no molecular‐based treatment recommendation based on a 90‐gene panel. Cancer Medicine 13(7): e7115.
- Todorova VK, Bauer MA, Azhar G, et al. (2024) RNA sequencing of formalin fixed paraffin-embedded heart tissue provides transcriptomic information about chemotherapy-induced cardiotoxicity. Pathology-Research and Practice 155309.
- Li J, Yan ZY, Liu WH, et al. (2024) Validation of TSO500 NGS panel for comprehensive genomic profiling from FFPE tumor samples. In: Proceedings of the American Association for Cancer Research Annual Meeting.
- Chen H, Liu Y, Wang F, Dingrong Zhong, Lingxiao Liu, et al. (2024) MeIS: DNA methylation-based immune response signatures for thyroid nodule diagnostics. The Journal of Clinical Endocrinology & Metabolism dgae141.
- Gargis AS, Kalman L, Bick DP, Justin M Zook, Ira M Lubin, et al. (2015) Good laboratory practice for clinical next-generation sequencing informatics pipelines. Nat Biotechnol. 33(7): 689-693.
- Rehm HL, Bale SJ, Bayrak-Toydemir P (2013) ACMG clinical laboratory standards for next-generation Sequencing. GENETICS in MEDICINE 15(9): 733-747.
- Cappello F, Angerilli V, Munari G, et al. (2022) FFPE-Based NGS Approaches into Clinical Practice: The Limits of Glory from a Pathologist Viewpoint. J Pers Med 12(5): 750.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.