Mini Review 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Learning to Discover: The Impact of AI in Preclinical Drug Development
*Corresponding author: Jonathan RT Lakey, Department of Surgery and Biomedical Engineering University of California Irvine.
Received: November 11, 2024; Published: November 15, 2024
DOI: 10.34297/AJBSR.2024.24.003245
Abstract
Drug discovery is the complex endeavor of identifying therapeutic compounds that are both safe and effective from a vast and ever-increasing chemical space. The high-throughput screening systems that have been used to identify hits have largely been replaced by rational drug discovery using expert-crafted machine learning algorithms to assist with hit identification, target prioritization, and lead optimization. However, persistently low clinical trial success rates continue to keep the cost of drug development high. This review outlines the history of machine learning in preclinical drug discovery of small molecules and highlights how parallel advancements in pharmacology, bioinformatics, and artificial intelligence (AI) have converged to create efficient AI-powered drug discovery tools. More specifically, biology has become digitized through modern methods for multi-omic data collection. Multi-omic data represents in-depth collections of biomarkers derived from DNA (genomics), RNA (transcriptomics), proteins (proteomics), metabolites (metabolomics) and lipids (lipidomics). These disparate datasets can be combined through neural networks and linked with hidden Markov models to create an interactome which attempts to mimic human systems biology within computer-based algorithms. Integration of transformer-powered deep learning models further improve these systems by overcoming the limitations of reductionist strategies for drug discovery. As customized, deep learning architectures become increasingly adopted, AI-driven predictive biology will continue to improve and soon become a mainstay in drug development.
Keywords: Deep learning, Transformers, Drug Discovery, Machine Learning, Multi-omics, Machine Learning, Bioinformatics
Introduction
The pharmaceutical industry has witnessed a persistent decline in productivity despite significant scientific and technological advancements during the seventy years. Since 1950, the number of new drugs approved by the FDA has been nearly cut in half every decade, resulting in an eighty-fold decline in inflation-adjusted efficiency. [1]. The average timeline from initial discovery to regulatory approval ranges between ten to fifteen years [2], with per-drug development costs ranging widely from $113 million to over $6 billion [3]. Scannell, et al. (2012) describe the troubling rise in drug development costs and declining returns as “Eroom's Law,” which is Moore's Law spelled backwards.
Amidst the challenges facing modern drug development, the pivotal question emerges of how artificial intelligence (AI) can help address the productivity problem within the pharmaceutical industry? To date, AI has been integrated within the target-based pharmacology paradigm through high throughput virtual screens, absorption, distribution, metabolism, and excretion (ADME) predictions, and in lead optimization. Better and smarter AI will undoubtedly continue to improve drug-target specificity predictions. However, these machine learning applications do not address the structural issues underlying Eroom’s Law, namely, that target-based approaches do not capture the complexity of biological systems and disease processes.
Productivity in the pharmaceutical industry has been a persistent challenge throughout the history of drug development. Drug discovery has evolved significantly over the past century, transitioning from a largely serendipitous, phenotype-based approach to a target-based paradigm due to advances in molecular biology and genomics that promised increased efficiency and specificity [4]. However, despite the dominance of target-based approaches, only 9.4% of clinically tested small-molecule drugs have been FDA approved using these newer methods [5]. This suggests that target-based strategies may not be as effective in yielding successful small molecule drugs as once hoped.
Phenotypic Drug Discovery
There has been a resurgence in phenotypic drug discovery (PDD) in recent years. PDD identifies potential therapeutic compounds based on observable change in cellular or organismal phenotypes without requiring prior knowledge of specific drug targets [6]. This approach has been instrumental in discovering drugs with novel or poorly understood mechanisms as PPD methods begin to ddress the profound complexity of biological systems.
Breakthroughs in the mid 20th century ranging from the elucidation of the structure of DNA to an improved understanding of drug-target interactions has catalyzed the shift from empirical drug discovery to target based approaches. The revival of phenotypic drug discovery is driven by the digitization of biological information and deep neural networks that can manage these extremely large datasets. Recent advancements in PDD have been propelled by the development of sophisticated in vitro models, high-content imaging technologies, and comprehensive mechanism-of-action profiling techniques [6]. Additionally, the integration of human based phenotypic platforms across the drug discovery pipeline enhances the process of hit triage, prioritization, and the elimination of compounds with undesirable mechanisms of action [7].
Despite these promising advancements, PDD faces challenges including efficient handling of data complexity and target deconvolution. The integration and analysis of massive biological datasets from high-content imaging and multi-omic technologies are difficult and time-consuming. Identifying the precise molecular targets of active compounds, a process known as target deconvolution, also remains a complex and expensive endeavor. However, these challenges can now be mitigated by leveraging the strengths of transformer-powered deep learning architecture.
AI Applications in Drug Discovery
Historically, neural networks were not the predominant machine learning (ML) technique in drug discovery like they are today. Instead, methods such as Support Vector Machines (SVMs) and decision trees were favored for their robustness when applied to compound classification, property prediction, and virtual screening [8]. SVMs are particularly adept at handling nonlinear problems and operate efficiently within high-dimensional feature spaces such as the vast chemical space [9] diagnostic image analysis in histology [10].
Decision trees have also been invaluable for data mining in hit discovery, drug metabolism, toxicology, and drug surveillance; they can handle diverse data formats and provide visual model representations, which facilitate the interpretation of complex biological data [11]. For virtual screening, decision trees excel in ligand-based approaches by delivering computationally efficient results that inform the chemical database queries and generate hypotheses about molecular actions.
Building upon the limitations of these foundational machine learning techniques, transformer-powered deep neural networks introduced a paradigm shift in searching the chemical space. The transformer architecture, originally developed for natural language processing (NLP), relies on attention mechanisms to process input data, addressing the parallelization problem of recurrent neural networks (RNNs) and long short-term memory networks (LSTMs) [12]. The self-attention mechanism enables capture of long-range dependencies within the data, a feature which renders neural networks applicable to a wide range of complex biological problems.
With modern AI tools, it has become feasible to build complex assays that leverage systems biology, capturing intricate properties through multiparametric readouts like gene expression profiles, protein interaction networks, metabolic flux analysis, and cellular phenotypic assays. Unlike traditional ML methods that often require extensive feature engineering and are limited in handling diverse and large-scale datasets, transformers excel in automatic feature extraction and integration of heterogeneous data sources [13]. This capability is particularly beneficial in drug discovery, where data types range from high-content imaging to multi-omic biomarker profiling and patient-derived metadata.
Furthermore, transformers facilitate the integration of multimodal data, combining information from various sources to provide a comprehensive view of biological systems through embeddings in a latent space. They can be applied to identify novel drug targets, predict compound efficacy, identify non-obvious phenotypic associations and assess potential toxicity with far greater accuracy. The versatility of transformers also extends their applications into generative tasks, for de novo design of new molecules with desired therapeutic properties [14].
AI-powered Drug Discovery
As an example of these developments from the authors’ laboratory, GATC Health employs a comprehensive AI-powered pipeline to enhance the preclinical drug discovery process (Figure 1). Assay development begins with a pretrained AI “head” that uses predictive modeling for selecting disease-relevant models. Following systems biology principles, these assays attempt to balance tractability and complexity by generating measurable, AI-deconvolvable results including gene expression profiles, protein interaction networks, metabolic flux analysis, and cellular phenotypic assays, while capturing the intricate interactions of biological systems. In the High-Throughput Phenotypic Screening step, AI-driven data analysis of assay data enables efficient identification of hit compounds for validation in in-vivo models. Mechanism of Action studies benefit from AI-informed target identification and pathway analysis and provide deeper insights into drug interactions and therapeutic mechanisms for rational drug design. During lead optimization, AI-guided molecular design facilitates the optimization of pharmacokinetic properties, considering factors such as bioavailability, off-target effects, mode of administration, and metabolic stability, for the development of robust drug candidates. Finally, the Investigation New Drug (IND) application process is streamlined through AI-supported regulatory compliance and preclinical safety testing, alongside the preparation of comprehensive clinical protocols.
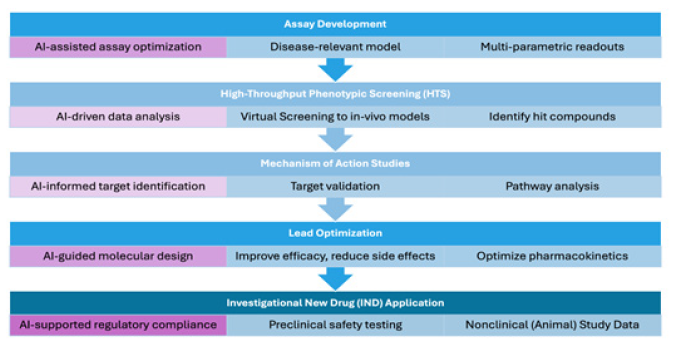
Figure 1: AI-Powered Phenotypic Drug Discovery Pipeline: Pretrained AI “Heads” streamlines preclinical drug discovery efforts.
The integration of AI into the drug discovery pipeline offers a solution to the persistent productivity challenges highlighted by Eroom’s Law. Leveraging the transformer’s ability to handle complex, multimodal biological data enables the design of phenotypic assays that better capture the intricacies of biological systems and disease processes. As AI becomes increasingly integrated throughout the entire process, the expectation is that this comprehensive application not only enhances the precision and efficiency of each stage but also accelerates timelines and reduces costs. Although challenges like interpretability and transparency remain, fully embracing AI-driven methodologies may ultimately serve to reintroduce serendipity to an otherwise highly regulated discipline.
References
- Scannell JW, Blanckley A, Boldon H, Warrington B (2012) Diagnosing the decline in pharmaceutical R&D efficiency. Nature reviews. Drug discovery 11(3): 191-200.
- Southey MW Y, Brunavs M (2023) Introduction to small molecule drug discovery and preclinical development.
- Rennane S, Baker L and Mulcahy A (2021) Estimating the Cost of Industry Investment in Drug Research and Development: A Review of Methods and Results. Inquiry: a journal of medical care organization, provision and financing 58: 469580211059731.
- Eder J, Herrling PL (2016) Trends in Modern Drug Discovery. Handbook of experimental pharmacology 232: 3-22.
- Sadri A (2023) Is Target-Based Drug Discovery Efficient? Discovery and "Off-Target" Mechanisms of All Drugs. Journal of medicinal chemistry 66(18): 12651-12677.
- Swinney DC and Lee JA (2020) Recent advances in phenotypic drug discovery. F1000Research, 9: F1000.
- Berg EL (2021) The future of phenotypic drug discovery. Cell chemical biology 28(3): 424-430.
- Heikamp K, Bajorath J (2014) Support vector machines for drug discovery. Expert opinion on drug discovery 9(1): 93-104.
- Simões RS, Maltarollo VG, Oliveira PR, Honorio KM (2018). Transfer and multi-task learning in QSAR modeling: Advances and challenges. 6(9): 74.
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, et al. (2023) Attention is all you need.
- Rodríguez Pérez R, Bajorath J (2022) Evolution of Support Vector Machine and Regression Modeling in Chemoinformatics and Drug Discovery. Journal of computer-aided molecular design 36(5): 355-362.
- Kremling H, Zunhammer F, Schütze K, Lernhardt W (2017) Cell Analysis Re-defined, Laser+Photonics. 60-63.
- Hammann F, Drewe J (2012) Decision tree models for data mining in hit discovery. Expert opinion on drug discovery 7(4): 341-352.
- Renqian Luo, Liai Sun, Yingce Xia, Tao Qin, Sheng Zhang, et al. (2022) BioGPT: generative pre-trained transformer for biomedical text generation and mining, Briefings in Bioinformatics 23(6).



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.