Review Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Multiomics Experimental Design for Drug Discovery
*Corresponding author: Jonathan Lakey, GATC Health Inc.2030 Main Suite 650, Irvine, CA 92614, Adjunct Professor, West Virginia University, Morgantown, WV 26506.
Received: January 19, 2025; Published: January 22, 2025
DOI: 10.34297/AJBSR.2025.25.003340
Abstract
The term “omics” refers to the branches of science that study the entirety of types of biomolecules present within living organisms including genomics; transcriptomics; proteomics and metabolomics, among others. Each “omic” layer uncovers a unique molecular story about a cell or tissue sample. For example, genomics informs what can happen, transcriptomics indicates what might happen, proteomics describes what makes it happen, and metabolomics reveals what is presently happening. While single-omic studies have been useful for identification of biomarkers, they lack the prognostic or predictive power needed to address the missing heritability problem, which manifests as three key genetic gaps: the numerical gap, the predictive gap, and the mechanistic gap. In contrast, a layered multi-omic approach offers the promise of true in silico modeling of biological systems which can predict perturbations and bridge the mechanistic gap by integrating diverse molecular layers to generate novel insights which individual omics approaches often miss. However, the integration of multi-omics data is complex and fraught with technical and computational challenges, particularly when combining vertical molecular layers with distinct parameters and statistical distributions. In addition, vertical integration exacerbates the concept of dimensionality (P≫N), this occurs when the number of features (P) far exceeds the number of samples (N), leading to over-training of algorithms and breakdown of statistical and machine learning models which have been optimized for sample-rich spaces. To address this issue, single-cell and spatial multi-omic studies should be included. Single-cell omics enable cellular level molecular resolution which can resolve heterogeneity and significantly increase sample numbers through cell-by-cell readouts. Spatial multi-omic analyses will preserve the spatial context of molecular data. Given the many considerations involved in multi-omic study designs, from data acquisition to feature analysis, this review aims to provide a comprehensive roadmap for experimental design with strategies to improve data integration which can assist in harnessing the power of systems biology for drug discovery.
Keywords: Deep learning, Transformers, Drug Discovery, Machine Learning, Multi-omics, Machine Learning, Bioinformatics
Introduction
The past century witnessed remarkable theoretical and technical advancements including the birth of molecular biology. The pioneering experiments of Hershey and Chase in 1952 demonstrated that DNA is the genetic material, while the structural elucidation of DNA by Crick, Watson, Franklin and Wilkins unveiled the iconic double-helix molecular structure. Building on these discoveries, Francis Crick proposed the central dogma of molecular biology in 1958, outlining the unidirectional flow of genetic information from DNA to RNA to proteins [1].
The sequencing of the human genome brought forth unexpected challenges and ultimately reshaped our understanding of genetic complexity. Two parallel efforts-the publicly funded International Human Genome Sequencing Consortium and the private company Celera Genomics-released their findings in Nature and Science, respectively, in 2001 [2-4]. These publications marked a monumental achievement in genomics, unveiling the draft human genome sequence and providing the first comprehensive map of human genetic material.
The genome draft revealed that humans have approximately 20,000 protein-coding genes, a finding that challenged long-standing assumptions about genome complexity. This discrepancy, referred to as the gene number paradox or G-value paradox, highlights the unexpected observation that humans, despite their complexity, possess a similar number of genes as simpler organisms, such as the nematode Caenorhabditis elegans (19,000 genes) or the mustard plant Arabidopsis thaliana (27,000 genes) [5]. The paradox underscored that organismal complexity is not solely dictated by the number of genes but also by the regulatory mechanisms, RNA-splicing and multiple interactions between genetic elements.
To address these questions, the ENCODE (Encyclopedia of DNA Elements) project was launched with the goal to identify and catalogue all known functional elements in the human genome [6]. One of the major findings from this work was that 80.4% of the human genome exhibits functionality in at least one cell type and revealed the critical roles of regulatory elements including enhancers, promoters, and insulators, all of which were formally classified simply as “junk DNA.” These results emphasize that biological complexity arises from dynamic regulatory networks, rather than simply from gene count.
Recognizing the new complexity of biological systems, researchers began to adopt systems biology methodologies to move beyond reductionist approaches. Systems biology involves generation of scale-free networks to quantitatively assess relationships between multiple intermediates within and across molecular pathways [7]. This integrative approach allows investigators to model the intricate web of molecular interactions driving biological processes, thus providing a more holistic understanding of cellular function. These revelations highlighted the need for multiomics, a systems- level strategy that integrates data from genomics, transcriptomics, proteomics, metabolomics, lipidomics and epigenomics. By bridging molecular layers, multiomics offers a powerful framework to connect genotype to phenotype and unravel the mechanisms underlying complex traits and diseases.
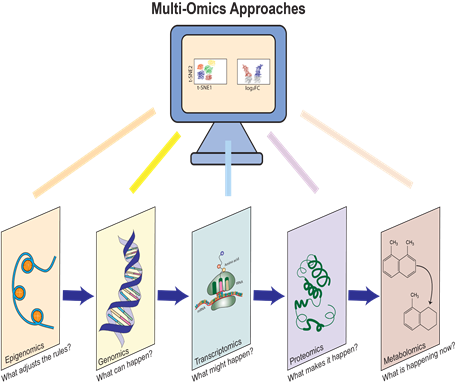
Figure 1 shows various omics layers, each accounting for its respective molecular role, following a logical progression from gene-level regulation to cellular function. Epigenomics defines gene regulatory rules, genomics outlines genetic potential, transcriptomics represents gene activity, proteomics describes functional machinery, and metabolomics captures dynamic biochemical processes which include subfields like lipidomics.
Each omics layer - epigenomics, genomics, transcriptomics, proteomics, and metabolomics - provides insight into a distinct molecular layer of the cell which can be integrated across multiple layers for a multiomics, systems level biology understanding (Figure 1).
Genomics (represented by the DNA double helix) deciphers the genetic blueprint, answering “what can happen” at the molecular level. Epigenomics (symbolized by the modifications on DNA) reveals regulatory mechanisms that modulate gene expression without altering the sequence. Transcriptomics (depicted by RNA and ribosomes) captures the dynamic activity of gene expression, showing “what might happen” by identifying active transcripts. Proteomics (illustrated by a folded protein structure) describes “what makes it happen,” highlighting the molecular machines that carry out cellular functions. Metabolomics (shown by small molecule structures) reflects the real-time state of cellular metabolism, answering “what is happening now” through the study of metabolic intermediates.
Genomics reveals the static blueprint of an organism’s genetic material, identifying variations in DNA sequences that underlie inherited traits and disease susceptibility. Next-generation sequencing (NGS) technologies have enabled rapid and precise detection of genomic mutations and novel biomarkers for precision medicine [8]. The genomic approach involves DNA extraction, amplification, and sequencing, followed by bioinformatics analysis for biomarker identification [9]. The human genome contains 4.1 to 5.0 million gene variants compared to the reference genome, with 99% arising from SNPs (Single Nucleotide Polymorphisms) and short indels [10].
Epigenomic studies catalog heritable changes in gene expression that do not alter the DNA sequence, including DNA methylation, histone modifications, chromatin structure, and non-coding RNAs [11,12]. These modifications regulate gene expression and mediate cellular responses to environmental factors (e.g. diet and stress) in a manner that expands the central dogma.
The focus of transcriptomics is on RNA transcripts including mRNA, lncRNA (long non-coding RNA) and microRNA, offering a snapshot of gene expression and regulation. High-throughput sequencing technologies, including RNA-seq, enable accurate transcript profiling and can resolve differentially expressed genes [13]. These methods, along with ChIP-seq, have advanced our understanding of signaling pathways and transcription factor regulation [13]. Genome-wide expression studies, employing technologies like microarrays and single-cell RNA-seq, can provide descriptive maps for both clinical and research applications [14].
Proteomics provides insight into the structure, abundance, and interactions of proteins, the molecular machines responsible for executing cellular functions. Advances in mass spectrometry-based proteomics have significantly expanded our understanding of protein- protein interactions (PPIs) and post-translational modifications (PTMs), which are critical for cellular function and signaling [15]. PTMs can trigger unique PPIs, such as with reader proteins, or indirectly alter interaction networks by inducing conformational changes or subcellular relocalization [16].
Metabolomics is the study of global metabolite profiles in biological systems and primarily uses advanced analytical techniques such as mass spectrometry and nuclear magnetic resonance to analyze thousands of small molecules in cells, tissues, or biological fluids [17,18]. Metabolomics captures small molecules and metabolites, providing a real-time view of cellular metabolism, with subfields such as lipidomics being commonly recognized. Metabolomics serves as a powerful tool for linking phenotypes to genotypes and understanding global systems biology [19].
Limitations of Single Omic Approaches
Single-omics technologies have revolutionized our understanding of biology by offering powerful tools to uncover key molecular mechanisms and disease-associated biomarkers. Genomics has identified genetic mutations responsible for heritable diseases like cystic fibrosis, sickle cell anemia, and Huntington’s disease, enabling the development of gene therapies [20,21]. Transcriptomics has advanced our understanding of gene expression and regulatory networks to enable antisense oligonucleotides therapy. Spinraza is a synthetic antisense oligonucleotide that modulates the splicing of the SMN2 gene, enabling it to produce full-length SMN protein, compensating for the loss of functional SMN1 protein in spinal muscular atrophy (SMA) [22]. Proteomics has revealed protein interactions, post-translational modifications, and their roles in cellular functions to inform drug efficacy and combination therapies such as in the mapping of the anaplastic lymphoma kinase (ALK) interactome for ceritinib, a drug for ALK-positive lung cancer [23]. Finally, metabolomic studies have identified key metabolic signatures, offering real-time insights into cellular metabolism.
Single-omics studies tend to focus on isolated aspects of molecular biology, overlooking the interconnective nature of biological processes. While genomics identify genetic variants, this approach does not reveal whether these variants are expressed or functionally relevant. Similarly, transcriptomic studies provide information on gene expression but often fail to correlate transcript levels with protein abundance or function. Proteomics offers a snapshot of protein abundance and activity but lacks upstream regulatory context or downstream effects on metabolites. Finally, metabolomics reflects metabolic changes but cannot elucidate the genetic or proteomic drivers of these changes.
A key challenge in this area is the missing heritability problem (MHP), which highlights the inability of single-omics approaches, such as genomics alone, to fully link genotype to phenotype [24,25]. Genome-wide association studies (GWAS) can identify numerous genetic variants associated with a trait, yet these variants explain only a small fraction of the heritable phenotypic variation. The limitations of single omic can be conceptualized as three key gaps. The numerical gap refers to the failure of single-omic studies to explain the full variance observed in traits or diseases. The prediction gap arises from the limited ability of single-omic approaches to build accurate, robust models for predicting disease onset, progression, or therapeutic response which is relevant for the development of better diagnostic and prognostic tools [25]. Finally, the mechanism gap highlights the challenge of elucidating causal relationships, as single-omics studies often reveal correlations without providing insight into the underlying biological mechanisms driving the observed phenotype which is relevant for developing new drug targets or exploring novel mechanisms of action [25].
Multiomics for a Systems Biology Understanding
Multiomics approaches, powered by advancements in technologies such as next-generation sequencing (NGS) and high-resolution mass spectrometry, have not only improved the numerical gap by generating vast amounts of data but are also addressing the prediction gap by uncovering complex molecular interactions and revealing patterns not discernible with single-omics approaches. Researchers have shown that as little as 10 microliters of capillary blood collected at home, can accurately profile proteins, metabolites, and lipids using mass spectrometry methods with very high correlation to omic data collected from traditional venous blood draws [26]. Developing surrogate models with lower-cost assays can make multi-omics approaches commercially feasible and scalable. Longitudinal studies combining transcriptomics, proteomics, and metabolomics have enabled the identification of predictive biomarkers for aging-related diseases and preterm birth risks, demonstrating the power of multiomics to bridge temporal gaps in disease progression [27]. In COVID-19, the horizontal integration of bulk metabolomics from serum, saliva, and sebum has revealed distinct metabolic profiles, identified key biomarkers, and highlighted significant pathway alterations. [28].
Multiomics integration has also bridged the mechanism gap by uncovering novel causal relationships. Integration of spatial transcriptomics and proteomics at the maternal-fetal interface revealed that fetal extravillous trophoblasts (EVTs) primarily drive the invasion of maternal tissue and remodeling of uterine spiral arteries, challenging previous assumptions that maternal immune cells were responsible. These findings offer key insights into placental development and implications for pregnancy complications, such as preeclampsia, where abnormal artery remodeling affects placental function and fetal growth [29]. In cancer, multiomics approaches have been instrumental in elucidating mechanisms of drug resistance. Studies on tamoxifen resistance in estrogen receptor- positive (ER+) breast cancer integrated transcriptomics and metabolomics to reveal lipid metabolic reprogramming and dysregulated ketogenesis as central drivers of resistance, identifying 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) synthase 2 as a potential therapeutic target [30,31].
Challenges of Multiomics Integration
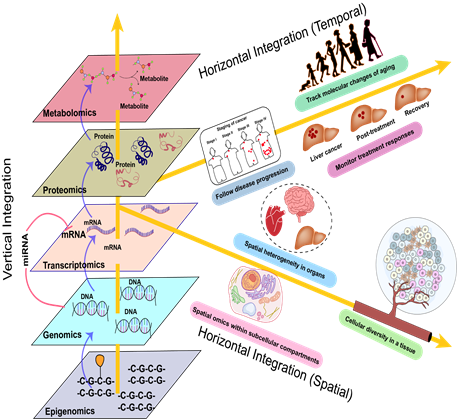
Successful multiomic experimental design involves structuring experiments to enable data integration either vertically and/ or horizontally (Figure 2). Vertical integration links different omics data types together (e.g. epigenomics, genomics, transcriptomics, proteomics, and metabolomics) [32]. While this approach offers a comprehensive view of molecular interactions across layers, it also exacerbates the challenge of dimensionality. Dimensionality issues arise when the number of features (P) far exceeds the number of samples (N), leading to sparse data distributions, reduced statistical power, and unreliable model predictions. This conundrum is particularly problematic for biological data, which are inherently high-dimensional, and becomes more pronounced as the integration of multiple omics layers significantly increases the number of features. Statistical and machine learning models optimized for sample-rich, lower-dimensional data, break down at this boundary, leading to overfitting and reduced model performance [33].
Horizontal integration aggregates the same omics data type across multiple studies or datasets within the same molecular layer [34]. This strategy is particularly valuable for integrating publicly available datasets, such as genomics repositories, or reconciling longitudinal data collected across different institutions. Addressing batch effects, or the technical variations between datasets that can obscure biological signals, is a well-characterized bioinformatics challenge with established solutions [35,36]. Effective batch effect correction is critical for harmonizing these datasets, ensuring that insights into temporal changes (e.g., disease progression or recovery) or spatial variability (e.g., tissue heterogeneity) remain robust and reproducible. Temporal (longitudinal) integration allows for the characterization of dynamic processes including aging, disease progression, and treatment responses while spatial integration enables the exploration of tissue heterogeneity, cellular diversity, and subcellular interactions. This approach is particularly valuable for uncovering the spatial organization of molecular pathways and their role in tissue- or cell-specific functions, as well as for identifying localized dysregulations in disease states (Figure 2).
Figure 2 illustrates two primary strategies for multiomics integration: vertical and horizontal. Vertical integration captures hierarchical relationships across molecular layers (epigenomics to metabolomics), showing how regulation flows from DNA modifications to metabolites. Horizontal integration explores temporal and spatial dimensions, tracking molecular changes over time (e.g., disease progression, treatment responses) and across spatial scales (e.g., tissue heterogeneity, cellular diversity).
To further tackle the dimensionality challenge, techniques like dimensionality reduction and intrinsic dimension analysis help transform data into lower-dimensional spaces without significant information loss [37]. Machine learning frameworks, such as OmiEmbed, also address this issue by effectively capturing patterns within high-dimensional datasets [38]. Single-cell multiomics significantly enhances the resolution of molecular studies by providing cell-by-cell readouts, addressing data sparsity, and resolving cellular heterogeneity. While single-cell approaches may initially be more expensive to run due to the complexity of data acquisition and processing, they can often be more cost-effective in the long term. Traditional methods require thousands to millions of bulk samples to achieve similar statistical power, which is impractical and prohibitively expensive, especially when each sample, such as a patient biopsy, can cost thousands of dollars depending on the type.
In contrast, single-cell multiomics treats each cell within a single sample as an individual data point, effectively multiplying the sample size and increasing statistical power without the need for additional physical samples. This not only reduces the cost per datapoint but also maximizes the value of rare and expensive samples, making single-cell approaches a practical and scalable solution for many applications to the extent that some would argue the most cost-effective strategy is to adopt single-cell approaches. However, single cell omics studies are not without its drawbacks. Single-cell methods often suffer from sparse data, with fewer reads per transcript in transcriptomic studies using platforms like 10x Genomics [38]. Sparsity makes it challenging to detect low-abundance proteins or RNA molecules, which require highly sensitive and finely tuned methods to capture accurately. These limitations underscore the importance of carefully considering whether to use bulk or single- cell approaches, as this choice represents a critical decision in multiomics experimental design (Figure 3). Each approach offers distinct advantages and drawbacks, and the decision should be guided by the specific research question, the type of biological insights sought, and the resources available.
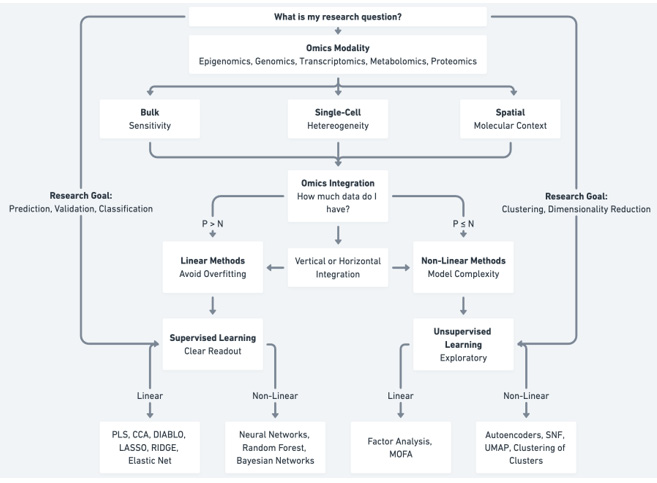
Figure 3: Decision tree for selecting omics modalities, integration strategies, and analytical methods.
Another challenge of multiomics integration is the specialized computational requirements of each omics layer and the sheer volume of data generated. While individual layers, such as genomics, transcriptomics, proteomics, and metabolomics, require tailored computational pipelines, understanding complex biological systems demands simultaneous analysis across all layers. This requires substantial computational power, interdisciplinary expertise, and advanced frameworks capable of synthesizing data into unified models. However, integration and data analysis often become bottlenecks, as well-established methods can generate a PhD’s worth of data in just a few weeks, yet the analytical capacity to process and visualize these big datasets still pose a challenge [39-41]. Addressing these issues remains critical to advancing multiomics research, emphasizing the need for innovative solutions in data integration and visualization to fully realize the potential of multiomic approaches.
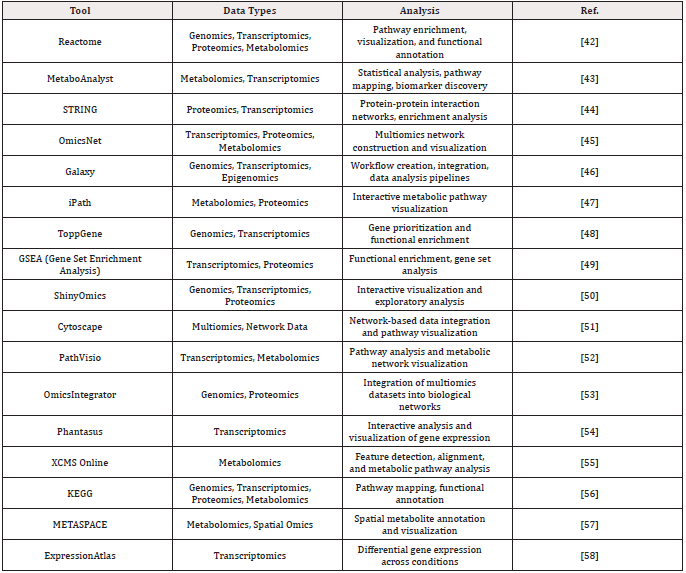
Web-hosted tools have emerged as critical resources to assist in providing accessible platforms for multiomics integration and analysis (Table 1). These tools streamline workflows by offering user- friendly interfaces, computational efficiency, and prebuilt pipelines for integrating diverse datasets, making them indispensable for researchers aiming to maximize the potential of multiomics approaches. Beyond simplifying analysis, web-hosted tools foster collaboration by enabling researchers across disciplines and institutions to share data, methodologies, and results in real time. Many of these tools support open-source frameworks, enabling the scientific community to contribute to the refinement and expansion of these platforms. This collaborative environment enhances the training of machine learning and deep learning models, as aggregated datasets from diverse users improve model robustness, accuracy, and generalizability.
Multiomics Experimental Design
The first and most critical step in designing a multiomics experiment is defining a clear and specific research question. A well-defined question provides the foundation for selecting the appropriate omics layers, integration strategies, and analytical methods. In the context of drug discovery, for example, the research question might be validating therapeutic targets while in personalized medicine, it would be designing a new prognostic tool using a surrogate predictive model distilled from amultiomics analysis. The more specific the question, the easier it becomes to tailor the experimental design to address it effectively (Figure 3).
In general, multiomics experiments can aim to predict biological outcomes, uncover novel mechanisms, or integrate diverse molecular layers to identify key biomarkers. Certain omics are more complementary than others and benefit from well-established workflows. For instance, genomics and epigenomics can be combined to study gene regulatory networks like in the case of integrating single cell ATAC-seq with single cell RNA-seq [59]. Transcriptomics and proteomics work together to connect gene expression with functional protein outputs [60]. Understanding these synergies enables the chosen omics layers to provide maximal insight while aligning with the overall research objectives.
The next key decision in multiomics experimental design is selecting between bulk, single-cell, or spatial approaches for each omics layer. This choice depends on the specific research objective, data requirements, and practical limitations, such as budget constraints or technical expertise. While there is often a technique available to achieve the desired resolution, specialized methods like single-cell metabolomics remain under development, with pipelines and protocols still evolving [61].
1. Bulk approaches are ideal when the goal is to capture overall averages and broad molecular trends across a sample. They are the most cost-effective and practical option, particularly when variations between individual cells or spatial context are not central to the research question [62].
2. Single-cell approaches are essential for studies focused on cellular heterogeneity, such as identifying rare cell populations, resolving dynamic cell states, or understanding diversity within tissues. These methods provide high-resolution insights but require greater expertise and resources [62-64].
3. Spatial approaches are necessary when the spatial organization of molecules, cells, or tissue architecture is critical. By preserving molecular context, these methods enable the study of localized interactions, such as those involved in development, disease progression, or tissue microenvironments [64,65].
Figure 3 outlines a structured flowchart for selecting omics modalities and analytical methods beginning with the choice of Bulk (sensitivity), Single-Cell (heterogeneity), or Spatial (molecular context) for a given omics datatype. For integration, data size (P: features; N: samples) determines the method: Linear Methods for P > N to avoid overfitting, leading to supervised learning for outcome prediction, and Non-Linear Methods for P ≤ N to handle model complexity, often in unsupervised learning for pattern discovery. Examples include linear (e.g., PLS, MOFA) and non-linear tools (e.g., Neural Networks, UMAP, clustering).
The next step in multiomics experimental design hinges on the dimensionality of the data, which plays a critical role in determining whether linear or nonlinear methods should be employed for analysis. Linear methods are often favored for their simplicity and ability to avoid overfitting, especially when the number of features is moderate, and the data allows for clear relationships to be captured without excessive model complexity. Conversely, nonlinear methods, such as those leveraging machine learning, excel when the relationships between variables are intricate and non-linear, provided that the model is robust [66].
The choice between these approaches becomes particularly relevant in the context of horizontal and vertical data integration. Vertical integration, which combines data from multiple omics layers (e.g., transcriptomics, proteomics, and metabolomics), typically increases the number of features in the dataset. This can skew the balance toward linear methods, as they are less prone to overfitting when handling large, feature-rich datasets. However, nonlinear methods remain preferable when the model incorporates accurate assumptions, adheres to best practices in machine learning such as avoiding data leakage or contamination and demonstrates minimal overfitting based on established metrics like cross-validation performance and independent test set evaluation [67,68].
For high-dimensional data (P > N), linear methods are recommended to minimize overfitting, with supervised learning approaches such as PLS (Partial Least Squares), CCA (Canonical Correlation Analysis), DIABLO (Data Integration Analysis for Biomarker discovery using Latent variable approaches for Omics studies), LASSO (Least Absolute Shrinkage and Selection Operator), and Elastic Net ensuring interpretable outputs (Figure 3). For complex, non-linear relationships, supervised techniques like neural networks, random forests, and Bayesian networks are applicable [40,69].
In contrast, for data where P ≤ N, non-linear methods can be employed to capture greater model complexity. Here, unsupervised learning facilitates exploratory analyses, with linear tools like Factor Analysis and MOFA (Multi-Omics Factor Analysis) or non-linear approaches such as autoencoders, SNF (Similarity Network Fusion), UMAP (Uniform Manifold Approximation and Projection), and clustering techniques revealing patterns and relationships [40,69].
The choice between supervised and unsupervised methods in multiomics analysis depends on the experimental goals. Supervised methods are generally employed when a clear readout, prediction, or validation of a hypothesis is required. For example, linear supervised methods, such as regression or classification models, are favored when relationships between features and the outcome are relatively straightforward and when avoiding overfitting is paramount. Nonlinear supervised methods, including machine learning approaches like decision trees or neural networks, are more suitable for capturing complex patterns in the data but require careful implementation to avoid overfitting and ensure generalizability [69,70].
In contrast, unsupervised methods are inherently exploratory, often serving as a first step in hypothesis generation or identifying underlying structures within the data. Linear unsupervised methods, such as principal component analysis (PCA), are ideal for reducing dimensionality and uncovering dominant trends in a dataset without imposing model complexity. Nonlinear unsupervised methods, such as t-SNE or UMAP, can reveal intricate patterns, clusters, or relationships that are not detectable through linear approaches [69,70]. However, these exploratory results often necessitate follow-up experiments or further validation to confirm biological relevance. Thus, the decision to use linear or nonlinear methods-whether supervised or unsupervised-should align closely with the specific aims of the study and the complexity of the data at hand.
Discussion
Multiomics represents a paradigm shift in biological research, offering a systems-level understanding of complex traits and diseases. Through the integration of diverse molecular datasets, multiomics transcends the constraints of single-omics approaches, offering profound insights into genotype-phenotype relationships and catalyzing advancements in drug discovery. Nevertheless, its full potential hinges on sustained innovation in experimental methodologies, integrative strategies, and computational frameworks.
The advent of the “whole data package” concept has emerged as a cornerstone in modern pharmaceutical research. In drug discovery, assembling a comprehensive dataset is often necessary to motivate stakeholders to pursue validation experiments. Isolated evidence from bulk sequencing or single-cell RNA-seq, for instance, may encounter skepticism regarding its reproducibility or biological significance. Critics might question whether such findings are substantiated by genetic evidence or merely reflect assay-specific artifacts. By synthesizing multiple data modalities-encompassing bulk, single-cell, and spatial omics-into a cohesive and robust package, researchers can mitigate these doubts and present a compelling case that resonates with seasoned drug discovery professionals. This integrative approach addresses the intrinsic limitations of individual datasets and cultivates confidence in the identification of novel therapeutic targets.
Convincing an audience of regulatory or scientific experts to embrace unproven targets is challenging. The “whole data package” paradigm, characterized by the synthesis of high-dimensional, multilayered datasets, establishes the requisite level of rigor to engender trust and enthusiasm among such stakeholders. This methodology ensures that candidate targets are scrutinized from diverse analytical perspectives, minimizing the risk of false positives and elucidating their biological relevance with unparalleled precision.
Looking forward, collaborative and interdisciplinary initiatives will be instrumental in unlocking the full potential of multiomics. The integration of artificial intelligence and machine learning into multiomics workflows holds immense promise for uncovering latent patterns and elucidating complex interdependencies within expansive datasets [71]. Simultaneously, the development of cost-effective and scalable techniques for single-cell and spatial omics will democratize access to these transformative tools, empowering researchers across both academic and industrial spheres to harness their capabilities in drug discovery.
By delivering a complete data package, multiomics has the potential to ameliorate the productivity problem in drug development. It streamlines the identification of therapeutic targets, reduces the time and cost associated with pre-clinical validation, and improves the predictive accuracy of drug efficacy and safety profiles. By interweaving high-dimensional datasets, multiomics unearths actionable biomarkers and mechanistic insights that elude traditional methodologies. Such a holistic perspective has the potential to revolutionize early-stage drug discovery, refine clinical trial design with precision, and expedite the transition of innovative therapies from bench to bedside.
References
- Lorentz, CP, Wieben ED, Tefferi A, Whiteman DA, Dewald GW (2002) Primer on medical genomics part I: History of genetics and sequencing of the human genome. Mayo Clinic proceedings 77(8): 773-782.
- Green ED, Chakravarti A (2001). The human genome sequence expedition: views from the "base camp". Genome research, 11(5), 645-651.
- Collins FS (2001) Contemplating the end of the beginning. Genome research 11(5): 641-643.
- Pennisi E (2001) The human genome. Science (New York, N.Y.) 291(5507): 1177-1180.
- Hahn MW, Wray GA (2002) The g-value paradox. Evolution & development 4(2): 73-75.
- Qu H, Fang X (2013) A brief review on the Human Encyclopedia of DNA Elements (ENCODE) project. Genomics, proteomics & bioinformatics 11(3): 135-141.
- Maron BA, Leopold JA (2016) Systems biology: An emerging strategy for discovering novel pathogenetic mechanisms that promote cardiovascular disease. Global cardiology science & practice (3): e201627.
- Yan J, Wang X (2020) CHAPTER 6. Detection of Disease-associated Mutations and Biomarkers Using Next-generation Sequencing.
- Alharbi RA (2022) Intersection of genomics and health informatics approaches in identification of diseases’ biomarkers. Journal of King Saud University - Science 34(1): 102264.
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al (2015) A global reference for human genetic variation. Nature 526(7571): 68-74.
- Gomase VS, Tagore S (2008) Epigenomics. Current drug metabolism 9(3): 232-237.
- Fingerman IM, McDaniel L, Zhang X, Ratzat W, Hassan T, et al. (2011) NCBI Epigenomics: a new public resource for exploring epigenomic data sets. Nucleic acids research, 39(Database issue) D908-D912.
- Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews. Genetics 10(1): 57-63.
- Tovar H, Alvarez Suarez DE, Gómez Romero L, Hernández Lemus E (2021) Bioinformatics of Genome-wide Expression Studies. Bioinformatics and Human Genomics Research.
- Pagel O, Loroch S, Sickmann A, Zahedi RP (2015) Current strategies and findings in clinically relevant post-translational modification-specific proteomics. Expert review of proteomics 12(3): 235-253.
- Wang S, Osgood AO, Chatterjee A (2022) Uncovering post-translational modification-associated protein-protein interactions. Current opinion in structural biology 74: 102352.
- Patti GJ, Yanes O, Siuzdak G (2012) Innovation: Metabolomics: the apogee of the omics trilogy. Nature reviews. Molecular cell biology 13(4): 263-269.
- Kaddurah Daouk R, Kristal BS, Weinshilboum RM (2008) Metabolomics: a global biochemical approach to drug response and disease. Annual review of pharmacology and toxicology 48: 653-683.
- Macel M, Van Dam NM, Keurentjes JJ (2010) Metabolomics: the chemistry between ecology and genetics. Molecular ecology resources 10(4): 583-593.
- Bloss C S, Jeste DV, Schork NJ (2011) Genomics for disease treatment and prevention. The Psychiatric clinics of North America 34(1): 147-166.
- Godbout K, Tremblay JP (2023) Prime Editing for Human Gene Therapy: Where Are We Now?. Cells 12(4): 536.
- Yang CW, Chen CL, Chou WC, Lin HC, Jong YJ, et al. (2016) An Integrative Transcriptomic Analysis for Identifying Novel Target Genes Corresponding to Severity Spectrum in Spinal Muscular Atrophy. PloS one, 11(6): e0157426.
- Haymond A, Davis JB, Espina V (2019) Proteomics for cancer drug design. Expert review of proteomics 16(8): 647-664.
- Young AI (2019) Solving the missing heritability problem. PLoS genetics 15(6): e1008222.
- Matthews LJ, Turkheimer E (2022) Three legs of the missing heritability problem. Studies in history and philosophy of science 93: 183-191.
- Shen X, Kellogg R, Panyard DJ, Bararpour N, Castillo KE, et al. (2024) Multi-omics microsampling for the profiling of lifestyle-associated changes in health. Nature biomedical engineering 8(1): 11-29.
- Jehan F, Sazawal S, Baqui AH, Nisar MI, Dhingra U, et al. (2020) Alliance for Maternal and Newborn Health Improvement, the Global Alliance to Prevent Prematurity and Stillbirth, and the Prematurity Research Center at Stanford University (2020). Multiomics Characterization of Preterm Birth in Low- and Middle-Income Countries. JAMA network open, 3(12): e2029655.
- Spick M, Lewis HM, Frampas CF, Longman K, Costa C, et al. (2022) An integrated analysis and comparison of serum, saliva and sebum for COVID-19 metabolomics. Scientific reports 12(1): 11867.
- Greenbaum S, Averbukh I, Soon E, Rizzuto G, Baranski A, Greenwald NF, (2023) A spatially resolved timeline of the human maternal-fetal interface. Nature 619(7970): 595-605.
- Hultsch S, Kankainen M, Paavolainen L, Kovanen RM, Ikonen E, et al. (2018) Association of tamoxifen resistance and lipid reprogramming in breast cancer. BMC cancer 18(1): 850.
- Hwang S, Park S, Kim JH, Bang SB, Kim HJ, et al. (2023) Targeting HMG-CoA synthase 2 suppresses tamoxifen-resistant breast cancer growth by augmenting mitochondrial oxidative stress-mediated cell death. Life sciences 328: 121827.
- Richardson S, Tseng GC, Sun W (2016) Statistical Methods in Integrative Genomics. Annual review of statistics and its application 3: 181-209.
- Gliozzo J, Soto Gomez M, Guarino V, Bonometti A, Cabri A, et al. (2025) Intrinsic-dimension analysis for guiding dimensionality reduction and data fusion in multi-omics data processing. Artificial Intelligence in Medicine 160: 103049.
- Richardson S, Tseng GC, Sun W (2016) Statistical Methods in Integrative Genomics. Annual review of statistics and its application 3: 181-209.
- Yu Y, Zhang N, Mai Y, Ren L, Chen Q, Cao Z, et al. (2023) Correcting batch effects in large-scale multiomics studies using a reference-material-based ratio method. Genome biology 24(1): 201.
- Ugidos M, Tarazona S, Prats Montalbán JM, Ferrer A, Conesa A (2020) MultiBaC: A strategy to remove batch effects between different omic data types. Statistical methods in medical research 29(10): 2851-2864.
- Mirza B, Wang W, Wang J, Choi H, Chung NC, et al. (2019) Machine Learning and Integrative Analysis of Biomedical Big Data. Genes 10(2): 87.
- Zhang X, Xing Y, Sun K, Guo Y (2021) OmiEmbed: A Unified Multi-Task Deep Learning Framework for Multi-Omics Data. Cancers 13(12): 3047.
- Liang A, Kong Y, Chen Z, Qiu Y, Wu Y, et al. (2023) Advancements and applications of single-cell multi-omics techniques in cancer research: Unveiling heterogeneity and paving the way for precision therapeutics. Biochemistry and biophysics reports 37: 101589.
- Rönn T, Perfilyev A, Oskolkov N, Ling C (2024) Predicting type 2 diabetes via machine learning integration of multiple omics from human pancreatic islets. Scientific reports 14(1): 14637.
- Subramanian I, Verma S, Kumar S, Jere A, Anamika K (2020) Multi-omics Data Integration, Interpretation, and Its Application. Bioinformatics and biology insights 14: 1177932219899051.
- Jassal B, Matthews L, Viteri G, Gong C, Lorente P, et al. (2020) The reactome pathway knowledgebase. Nucleic acids research, 48(D1): D498-D503.
- Xia J, Wishart DS (2011) Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nature protocols 6(6): 743-760.
- Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, et al. (2023) The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic acids research 51(D1): D638-D646.
- Zhou G, Xia J (2018). OmicsNet: a web-based tool for creation and visual analysis of biological networks in 3D space. Nucleic acids research 46(W1): W514-W522.
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, et al. (2018) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic acids research 46(W1): W537-W544.
- Letunic I, Yamada T, Kanehisa M, Bork P (2008) iPath: interactive exploration of biochemical pathways and networks. Trends in biochemical sciences 33(3): 101-103.
- Chen J, Bardes EE, Aronow BJ, Jegga AG (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic acids research 37(Web Server issue): W305-W311.
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102(43): 15545-15550.
- Surujon D, van Opijnen T (2020) ShinyOmics: collaborative exploration of omics-data. BMC bioinformatics 21(1): 22.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13(11): 2498-2504.
- Kutmon M, van Iersel MP, Bohler A, Kelder T, Nunes N, et al. (2015) PathVisio 3: an extendable pathway analysis toolbox. PLoS computational biology 11(2): e1004085.
- Tuncbag N, Gosline SJ, Kedaigle A, Soltis AR, Gitter A, et al. (2016) Network-Based Interpretation of Diverse High-Throughput Datasets through the Omics Integrator Software Package. PLoS computational biology 12(4): e1004879.
- Kleverov M, Zenkova D, Kamenev V, Sablina M, Artyomov MN et al. (2024) Phantasus, a web application for visual and interactive gene expression analysis. eLife, 13: e85722.
- Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G (2012) XCMS Online: a web-based platform to process untargeted metabolomic data. Analytical chemistry 84(11): 5035-5039.
- Kanehisa M, Furumichi M, Sato Y, Ishiguro Watanabe M, Tanabe M (2021) KEGG: integrating viruses and cellular organisms. Nucleic acids research 49(D1): D545-D551.
- Wadie B, Stuart L, Rath CM, Drotleff B, Mamedov S, et al. (2024) METASPACE-ML: Context-specific metabolite annotation for imaging mass spectrometry using machine learning. Nature communications, 15(1): 9110.
- Papatheodorou I, Moreno P, Manning J, Fuentes AM, George N, et al. (2020) Expression Atlas update: from tissues to single cells. Nucleic acids research 48(D1): D77-D83.
- Jansen C, Ramirez RN, El Ali NC, Gomez Cabrero D, et al. (2019) Building gene regulatory networks from scATAC-seq and scRNA-seq using Linked Self Organizing Maps. PLoS computational biology 15(11): e1006555.
- Du Y, Clair GC, Al Alam D, Danopoulos S, Schnell D, et al. (2019) Integration of transcriptomic and proteomic data identifies biological functions in cell populations from human infant lung. American journal of physiology. Lung cellular and molecular physiology 317(3): L347-L360.
- Lanekoff I, Sharma VV, Marques C (2022) Single-cell metabolomics: where are we and where are we going?. Current opinion in biotechnology 75: 102693.
- Li Y, Ma L, Wu D, Chen G (2021) Advances in bulk and single-cell multi-omics approaches for systems biology and precision medicine. Briefings in bioinformatics 22(5): bbab024.
- Lim J, Park C, Kim M, Kim H, Kim J, et al (2024) Advances in single-cell omics and multiomics for high-resolution molecular profiling. Experimental & molecular medicine 56(3): 515-526.
- Vandereyken K, Sifrim A, Thienpont B, Voet T (2023) Methods and applications for single-cell and spatial multi-omics. Nature reviews. Genetics 24(8): 494-515.
- Zhou R, Yang G, Zhang Y, Wang Y (2023) Spatial transcriptomics in development and disease. Molecular biomedicine 4(1): 32.
- Makrodimitris S, Pronk B, Abdelaal T, Reinders M (2023) An in-depth comparison of linear and non-linear joint embedding methods for bulk and single-cell multi-omics. Briefings in bioinformatics 25(1): bbad416.
- Schultz BG, Joukhadar Z, Nattala U, Quiroga M del M, Bolk F, et al. (2021) Best practices for supervised machine learning when examining biomarkers in clinical populations. In A. A. Moustafa (Ed.), Big Data in Psychiatry & Neurology. 1-34.
- Chen V, Yang M, Cui W, Kim JS, Talwalkar A, et al. (2024) Applying interpretable machine learning in computational biology-pitfalls, recommendations and opportunities for new developments. Nature methods 21(8): 1454-1461.
- Jovic D, Liang X, Zeng H, Lin L, Xu F (2022) Single-cell RNA sequencing technologies and applications: A brief overview. Clinical and translational medicine 12(3): e694.
- Feldner Busztin D, Firbas Nisantzis P, Edmunds SJ, Boza G, Racimo F, et al. (2023) Dealing with dimensionality: the application of machine learning to multi-omics data. Bioinformatics (Oxford, England) 39(2): btad021.
- Samuel Kho, Waldemar Lernhardt, Eric J Mathur, Jayson, Uffens, and Jonathan RT Lakey, et al. (2024) Learning to Discover: The Impact of AI in Preclinical Drug Development. Am J Biomed Sci & Res 24(5): 577-579.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.