Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Detection of Canine Parvovirus in Dogs by Means Polymerase Chain Reaction
*Corresponding author: Navarro C, Department of Animal Preventive Medicine, FAVET, University of Chile, Chile.
Received: February 26, 2020; Published: March 06, 2020
DOI: 10.34297/AJBSR.2020.07.001219
Abstract
Worldwide, the infection with canine parvovirus type 2 (CPV-2) is one of the main causes of hemorrhagic enteritis in dogs and having a diagnostic technique that is highly sensitive is essential for veterinarians, dog owners and breeders. In this work, a protocol was implemented that uses the conventional Polymerase Chain Reaction (PCR) to detect a CPV-2 DNA fragment from feces of dogs with clinical signology corresponding to canine parvovirus. In total, 12 stool samples that were positive with conventional PCR were collected and analyzed, which was confirmed by sequencing the fragments obtained and contrasted with the sequences of the different variants of CPV-2 described in the GenBank database.
The same samples were analyzed with a rapid test, which corresponds to a routine Immunochromatography (IC) technique in the veterinary practice. In this case of the 12 samples analyzed, only 41.7% were positive, showing a lower sensitivity than the molecular technique for the diagnosis of canine parvovirus. Additionally, an analysis of the nucleotide sequences obtained was made, yielding an average variability of 0.7%. The results of this work allow to establish that conventional PCR is a recommended diagnostic technique for the detection of canine parvovirus, but not the rapid tests used in the veterinary practice that in this and other studies have consistently shown low sensitivity. It is important to note that the present work is the first molecular approach to canine parvovirus type 2 in Chile.
Keywords: Canine Parvovirus; Polymerase Chain Reaction; Immunochromatography
Introduction
Canine Parvovirus type 2 (CPV-2) infection is one of the main causes of severe gastroenteritis in the canine species around the world. The virus that was detected in the late 1970s and identified in the early 1980s, has become a major threat to the tenure and breeding of dogs, since the infection has a morbidity that can reach up to 100% and a mortality that in puppies can be 90% [1] . The first cases of canine parvovirus in Chile were observed in 1980 and in 1981, the virus was isolated for the first time in the country [2] . Currently there are three variants of CPV-2 that infect dogs in the world and that have completely replaced the original CPV-2. These variants are called CPV-2a, CPV-2b and CPV-2c and differ only in some amino acids of the proteins that form their capsid.
These differences do not translate into large changes in the form of presentation of the disease, in the antigenic properties of the virus or in the efficacy of the vaccines available for active immunization in dogs. The importance of the infection and its implications for dog owners, breeders and veterinarians, makes it necessary to have an effective diagnostic tool, both to establish an adequate treatment in the patient, and take control measures to prevent the spread of the virus. With this background, in this work the implementation of a protocol of a conventional Polymerase Chain Reaction (PCR) technique was proposed, as a method for the detection of canine parvovirus, in samples from the depositions of sick dogs.
Background
In 1967, the first parvovirus that infects canines was detected, initially known as canine parvovirus, and which is now known as Canine Diminute Virus [3, 4]. Canine Parvovirus type 2, was detected later in 1978, after emerging as a pandemic due to lack of previous immunity in dogs, which allowed the rapid spread of the virus throughout the world [5, 6]. CPV-2 is the virus that is currently important, since its appearance, it became an important cause of morbidity and mortality in dogs around the world [3]. The origin of the virus is not clear, but it is postulated that it emerged as a variant of the Feline Panleukopenia Virus (FPV). Among other hypotheses about the emergence of this virus, it is postulated that it would be a mutation of the FPV present in the vaccine or an adaptation of the parvovirus that affect wild carnivores such as foxes and mink [7].
Although the CPV-2 virus was detected in 1978, studies suggest that it would have emerged in the early 1970s [8]. The detected CPV- 2 continued to evolve and in 1980 its first variant was identified, called CPV-2a, a variant that had substitutions in some amino acids of the sequence that forms the VP2 protein of the viral capsid. In 1984 a new variant was detected, this time from CPV-2a and that was called CPV-2b. Both spread rapidly around the world and in a short time completely displaced the original virus (CPV-2) [4]. In 2000, a new variant was described in Italy from CPV-2b, which was called CPV-2c, and which was also quickly detected in other parts of the world [4, 9].
Taxonomy, Structure and Viral Replication
The CPV-2 is classified within the family Parvoviridae, subfamily Parvovirinae [10] and was recently included in the genus Protoparvovirus, as a member of the species Protoparvovirus carnivore 1 which also includes, among others, FPV [11, 12]. Parvoviruses are small viruses, approximately 25nm in diameter, without sheath, with an icosahedral capsid composed of three structural proteins (VP1, VP2 and VP3), and a single-stranded DNA genome of approximately 5,000 nucleotides. This thread it encodes two structural proteins (VP1 and VP2) and two non-structural proteins (NS1 and NS2) [10]. The viral capsid is composed of 60 protein subunits (capsomeres), which is constituted by 90% by the VP2 protein and approximately 10% by the VP1 protein. Additionally, a third structural protein, VP3, can be found, which is not detected in all viruses, and which is originated by the action of a host protease on VP2 [13]. This protein is only present in the capsid when it contains the viral DNA [4].
The NS1 and NS2 proteins have roles during viral replication. NS1 has functions of helicase and endonuclease and interferes with the replication of host DNA, keeping the cell in the S phase of the cell cycle and NS2 regulates the expression of viral genes [13]. Because the parvovirus genome is small and only encodes some proteins, these viruses depend heavily on the host cell for replication. The proteins necessary for viral replication are found only in the S phase of the cell cycle [14], so replication occurs in the nucleus of cells in constant division, such as fetal cells, newborn or of intestinal tissue of young or adult animals [10]. By the action of DNA polymerase, the single strand of DNA of the virus is converted into a double strand. Two classes of messenger RNA (mRNA) are generated by cellular RNA polymerase II, one of which is longer than that encoded by nonstructural proteins and the other of shorter length that encodes the capsid proteins [14].
Pathogenesis
CPV-2 enters the body via the oropharynx [13], by contact with infected feces or contaminated surfaces [4]. The virus replicates initially in the lymphoid tissue of the region [15], in the mesenteric lymph nodes and in the thymus, and then spreads through the blood to the epithelium of the crypts in the mucosa of the small intestine [3], after four days from infection [15]. CPV-2 directly affects the cells of the intestinal crypts, leading to the destruction and shortening of the intestinal villi, which prevents the absorption of nutrients resulting in diarrhea. The deterioration of the intestinal mucosa allows the exit of blood towards the intestinal lumen and the passage of bacteria from the intestine to the blood [4]. The lymphoid tissue is also affected, and the destruction of lymphocytes will produce immunosuppression, predisposing to secondary infections[16]. The virus rarely affects the myocardium, and this occurs when the mother does not have antibodies [15] and the puppy acquires the infection within the first week of life, when the myocardial cells are in rapid division [13].
Clinical Signs
The enteric infection presents with anorexia, depression, vomiting, abdominal pain and eventually fever. Diarrhea can be severe and hemorrhagic, especially in puppies. Due to diarrhea and vomiting, dehydration occurs rapidly. Symptoms appear three to five days after the virus enters the body and the death of the puppy can occur three days after the onset of clinical signs [3, 4, 16, 17]. When CVP-2 infects the myocardium, clinical signs are usually evident throughout the litter [17] and death occurs due to congestive heart failure. 70% of puppies will die within the first eight weeks and 30% will suffer pathological changes in the organ, which will lead to death weeks or months later [16].
Treatment and Prevention
The treatment of the infection by CPV is like the one administered for other gastroenteritis, mainly of support and whose intensity will depend on the severity of the signs and symptoms. Initial consideration should be given to food restriction and parenteral nutrition in cases of persistent vomiting, the use of antiemetics and antidiarrheals, if necessary, and eventually the transfusion of whole blood or plasma in case of severe anemia [18].
Immunization
Puppies acquire antibodies through colostrum, which gives them protection against the virus in the first weeks of life. The highest rates of infection are observed in puppies older than six weeks of age. These maternal antibodies interfere with the immunization of puppies through vaccination. Titers of maternal antibodies equal to or greater than 1:80 given immunity to puppies. When the title is 1:40, it does not confer immunity, but it is capable of interfering with the active immunization of puppies [19]. Active immunization is done through the administration of vaccines and 90% of puppies respond to it at twelve weeks of age, when maternal immunity has declined. Generally, polyvalent vaccines are used that, in addition to CPV, contain among other antigens, canine distemper and leptospira. The vaccines commercialized in Chile and registered in the Agricultural and Livestock Service (SAG) correspond to polyvalent modified live virus vaccines [4, 20].
Diagnosis
Diagnosis
The diagnosis through clinical signs is only presumptive and must be confirmed by a diagnostic test. The tests used to detect the virus or viral antigen include hemagglutination, which despite being quick and simple to detect parvovirus in feces using porcine, feline or Rhesus monkey erythrocytes [4], is less sensitive than other tests such as viral isolation or the Polymerase Chain Reaction (PCR) [21], in addition to which some variants of CPV-2 would lack hemagglutinating activity [22]. A variation of this test is the inhibition of hemagglutination, which would be more specific, since specific antibodies against the viral antigen are used [21]. The ELISA test is fast and simple, but a great variability in its sensitivity has been found in several studies, 81.8% [23], 56.2% [21] and 18.4% [24], although it can detect the three circulating CPV-2 variants.It is also possible to perform electron microscopy, although CPV-1 and CPV-2 are morphologically identical [16], which does not allow their identification. The IC immunochromatography technique is routinely used in clinical practice, since it delivers the result in a short time, however, compared to molecular tests, its sensitivity does not exceed 50% [10]. PCR has proved to be the most sensitive test for the detection of canine parvovirus and, in its conventional form, has reached a sensitivity of 93.15%, compared with the real-time PCR technique as a reference test [21]. This test has been transformed 5 in the technique of choice against cases of dogs with clinical signs that are negative for other diagnostic tests [15]. Consequently, the purpose of this Title Report was to implement a molecular diagnostic technique of CPV-2, as is a conventional PCR technique, which amplifies a fragment of the gene that encodes the structural proteins of the viral capsid.
Materials and Methods
The present report was carried out in the laboratory of enteric viruses, Virology Program, ICBM, of the Faculty of Medicine of the University of Chile. As a positive control for the PCR reaction, viral DNA was obtained from the commercial vaccine NOBIVAC PUPPY DP®. This vaccine contains live virus modified canine parvovirus strain 154 (≥107.0 TCDI50) and canine distemper virus Onderstepoort strain (≥105.0 TCDI50). As a negative control, DNA from virus available in the laboratory (adenovirus and polyomavirus) was used. For the control of reagents, nuclease-free water was used.
Samples
The PCR technique implemented was evaluated in twelve samples of feces of dogs with compatible signology with canine parvovirus collected in different veterinary clinics of Santiago whose diagnosis was previously confirmed by an HF test. The stool samples were collected and stored at 4°C until the time of viral DNA extraction (Table 1).
DNA Extraction
Extraction columns (High Pure PCR Template Preparation Kit from Roche®) were used according to the manufacturer’s instructions. Briefly, 200μL of the sample (reconstituted vaccine or bloody diarrhea samples) were incubated in a 200μL solution of Binding buffer plus 40μL Proteinase K for 10 minutes at 72°C. After incubation, 100μL of Binding buffer was added and transferred to the column where it was centrifuged at 13,000 rpm for 1 minute. Then 500μL of Inhibitor Removal buffer was added to the column and centrifuged again at 13,000rpm for 1 minute. Afterwards, two washes were carried out with 450μL of Wash buffer. Finally, to the column already washed, 50μL of Elution buffer was added to recover the DNA already purified.
Primers
The primers used were designed by [9, 12, 21] in different studies. These primers amplify a DNA fragment of approximately 580 base pairs, which encodes the viral capsid proteins and encompasses the amino acids that determine the important biological properties of the virus [9]: 555forw 5 ‘CAGGAAGATATCCAGAAGGA 3’ and 555rev 5’ GGTGCTAGTTGA TATGTAATAAACA 3’. The primers were synthesized by the company IDT® (USA).
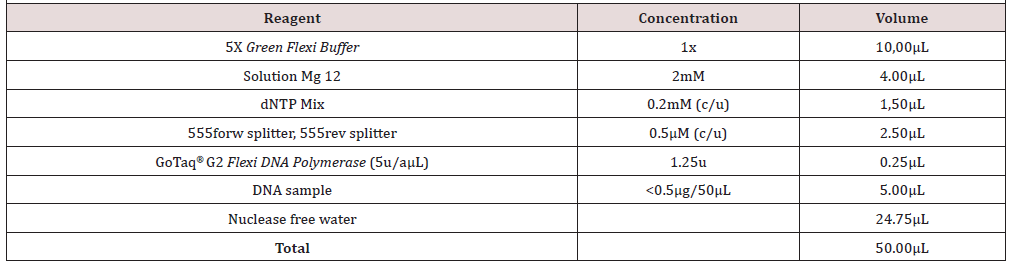
PCR Reaction Mixture
For the PCR reaction mixture, the GoTaq® G2 Flexi DNA polymerase commercial kit (Promega®) was used according to the manufacturer’s instructions (Table 2). PCR reaction. The PCR reaction was carried out in a 96-well thermal cycler (Applied Biosystems 2720, USA) of 0.2mL using the following protocol: activation of the Taq DNA polymerase at 94°C for 2 minutes. Subsequently, 40 denaturation cycles were carried out at 94°C for 30 seconds: Alignment at 48°C for 30 seconds and polymerization at 72°C for 1 minute and finally a final extension stage at 72°C for 10 minutes. 4.7 Display of the amplified product to visualize the product of the PCR reaction, a 2% agarose gel electrophoresis was performed in TAE buffer (Tris-Acetate-EDTA). The electrophoresis was performed at 90V for 40min, using a molecular standard as a molecular weight marker with fragments between 100 and 1000bp (BioLabs®). Subsequently the agarose gel was incubated in a GelRed™ solution (GelRed™ Nucleic Acid Gel Stain, Biotium, USA) for 30 minutes at room temperature. Then the gel was observed in an ultraviolet light transilluminator (UVP® Transilluminator) and photographed to record the results.
Determination of the identity of the amplified fragment. To determine the identity of the amplified fragment, after recording the results of electrophoresis in 2% agarose gel, the amplified fragment was extracted from the gel using the E.Z.N.A. Gel Extraction Kit (Omega Bio-tek®) according to the manufacturer’s instructions. In summary, the gel was cut to the size of the fluorescent band containing the desired PCR product. To this gel fragment an equal volume of Binding buffer was added and incubated for 7 minutes at 60°C. Then the content was transferred to an extraction column where it was centrifuged at 10,000rpm for 1 minute. The filtrate was discarded and 300μL of Binding buffer was added by centrifuging again. Subsequently, a wash was carried out with 700μL of Wash buffer. Finally, 50μL of Elution buffer was used to collect the desired fragment.
The extracts were stored at -20°C until they were sequenced in the company Macrogen® (Republic of Korea). To determine the percentage of nucleotide identity, the obtained sequences were entered into the BLAST™ online program (National Library of Medicine, USA), where the latter made alignments with the sequences found in its database, delivering those sequences with which produce meaningful alignments.
Analysis of Results
Those samples that generated an amplicon of approximately 600bp with a Nucleotide Identity Percentage (NIP) greater than 90% were considered positive.
Immunochromatography Technique (IC)
A commercial CI test was used to confirm the diagnosis of CPV-2 infection in each of the collected samples (SensPERT™ CPV AG TEST KIT, Vet All Laboratories, Republic of Korea) and was used according to the manufacturer’s instructions. The procedure consists of collecting around 30mg of stool with the torula included in the kit and homogenizing in the buffer also included. Once the fecal particles have been decanted, about 100μL of the supernatant is used, which is deposited in the reaction device, where it migrates by capillarity through a nitrocellulose membrane, and is conjugated with a first antibody against the antigen to be detected and a detection reagent. During the migration, it first arrives at a capture American Journal of Biomedical Science & Research 544 zone possessing a second antibody against the desired antigen that retains the antibody antigen conjugate producing the color change of the detection band of the device. There is another band that has an antibody that recognizes the detection reagent and captures the conjugate that is not retained in the capture zone. This line is the control of the test and its coloration indicates that the test was well performed.
Biosafety
In the laboratory there is limited access to the facilities, use of clean apron, use of clean material and proper disposal of waste. Regarding the PCR procedure and subsequent electrophoresis, nitrile gloves were used, both to avoid contamination of the sample, and to manipulate the gel. When using the transilluminator you must have glasses with UV filter and an acrylic plate located between the equipment and the person who visualizes the gel. Finally, the gel and gloves used were disposed of properly.
Results
Implement a PCR assay for the molecular diagnosis of canine parvovirus type 2. The observation of the 2% agarose gel in the transilluminator allowed the detection of bands or amplified close to 600bp, both in the positive controls and in the twelve samples analyzed. In the negative control and the reagent control, the presence of bands or amplicons (Figure 1).
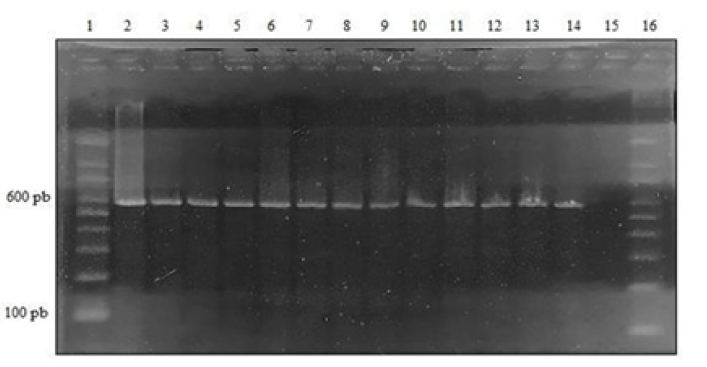
Figure 1: 2% PCR agarose gel CPV-2, lane 1: molecular size marker (100 to 1000bp); Lane 2: Positive Control (Novivac Puppy DP® Vaccine); Lane 3: Sample 1; Lane 4: Sample 2; Lane 5: Sample 3; Lane 6: Sample 4; Lane 7: Sample 5; lane 8: Sample 6; Lane 9: Sample 7; Lane 10: Sample 8; Lane 11: Sample 9; lane 12: Sample 10; Lane 13: Sample 11; Lane 14: Sample 12; lane 15: Reagent Control; Lane 16: molecular size marker (100 to 1000bp).
Determination of Nucleotide Identity
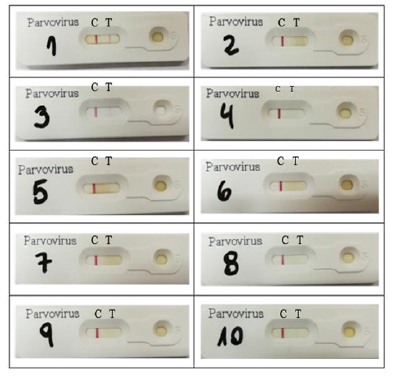
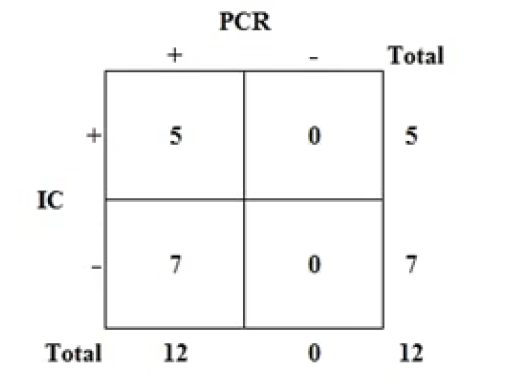
Figure 2: Results of the IC tests performed in the laboratory of enteric viruses. The red line in C indicates that the technique was well performed. Red line in T indicates positive test result. Samples 11 and 12 were analyzed by the same technique in the veterinary practice and were positive.
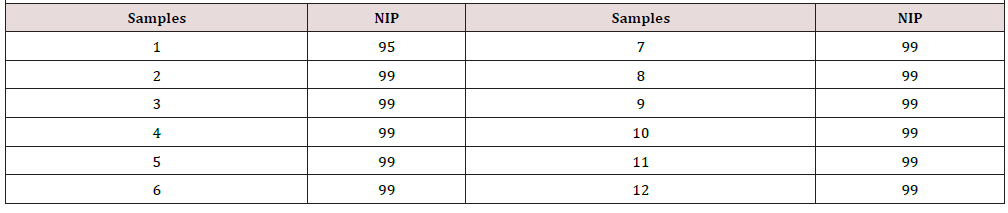
Table 3: Percentage of nucleotide identity with respect to the sequences of the different variants of CPV-2 described in the BLAST ™.
The sequences of the twelve samples that were analyzed were entered the BLAST™ program, free online access, generating significant alignments with different strains of the CPV-2 variants and delivering a percentage of nucleotide identity greater than 90% (Table 3) with respect to the canine parvovirus sequences described in the BLAST™ (Annex 1 and 2). To evaluate the PCR technique in stool samples of dogs with confirmed parvovirus by an immunochromatography technique. Thus, in the laboratory of enteric viruses, IC tests were performed on ten of the twelve samples obtained (Figure 2), the two remaining samples underwent IC directly in the veterinary practice, resulting positive. When the test was carried out in the laboratory, only three positive samples were obtained. In total, 5 positive samples were obtained by IC, that is, 41.7%. No invalid results were obtained.
Discussion
Currently, CPV-2 infection is one of the most frequent causes of infectious enteritis in dogs around the world. This disease has a high morbidity and mortality especially in puppies, which makes it a constant threat to the tenure and breeding of dogs. This makes it necessary to have an efficient diagnostic technique, not only to establish an adequate treatment for dogs that have become ill, but also to prevent the spread of the disease. Among the molecular diagnostic methods, conventional PCR is presented as a good alternative of choice because of its high sensitivity and high specificity, compared to traditional methods used clinically, such as the HF test. One of the findings of this work was the difference between the results of conventional PCR and HF. While the PCR detected 100% of the samples as positive, confirmed by the sequencing of the obtained amplicons, the CI was able to detect only 41.7% of the samples as positive for CPV-2. These results, represented in Figure 3, are conditioned by several studies that have determined that traditional diagnostic methods, such as HF, have lower sensitivity than molecular methods such as the conventional PCR used in this study [21, 24].
Comparison of the PCR with the CI (Figure 3). The IC shows a sensitivity of 41.7% (5/12) with respect to the conventional PCR. Some studies that have compared the performance of HF compared to conventional PCR, showed percentages of HF sensitivity like those achieved by PCR. [12, 25], described sensitivities of 72.22% and 84% respectively for the CI and recommend it as a diagnostic tool for routine use in the veterinary clinic mainly because of its low cost, the speed with which it delivers the result and its ease of application. However, the consensus is that the sensitivity of traditional diagnostic tests does not exceed 50% with respect to molecular methods in the detection of CPV-2 [10]. The results of this work are in line with the consensus.
A possible explanation to the result of this work and to the variability described in the sensitivity in the different studies that compare HF with conventional PCR, which may be 15.8% as reported by [247] or 84% reported by [27], it would be the existence of several factors that affect the antibody antigen reaction on which the CI technique is based. Among them, the main factor is the high concentration of viral antigen that is required to produce a visible band in the reaction device. Other factors such as the presence of water, blood or other fluids in the sample, and environmental humidity, to which the nitrocellulose membrane of the device is highly sensitive, can affect the migration of the antigen-antibody complex [26]. Additionally, the interpretation of the result also depends on the operator’s ability to detect the color change in the reaction device [27]. The presence of antibodies in the intestinal lumen would also decrease the number of viral particles available in the stool sample to perform the IC test [10].
On the other hand, the variant of the virus present in the sample would not affect the sensitivity of the diagnostic test, since all the variants are detected by this technique [28]. All the factors described could explain the obtaining of false negative results with HF and the different intensity in the color change of the samples that were positive. Although Real-Time PCR Has Advantages Over Conventional PCR, such as: Requiring less time in the processing of the samples, being more sensitive and at the same time allowing the quantification of the viral DNA of the sample, the first one requires more expensive equipment, more specialized reagents, such as the probe that allows the quantification of the amplified product [27], and more qualified personnel, which It becomes a test that is not applicable to veterinary practice routinely [21].
Regarding the primers used in this work, it should be noted that with these it is possible to detect any of the three variants of CPV-2, because they amplify a fragment of the gene that encodes the structural proteins and that encompasses the amino acids that provide the important biological properties of the virus [9]. Although it is possible to design specific primers for each variant of the virus [29], the mere presence of CPV-2, in any of its variants, is relevant in clinical practice, since it has not been observed a significant difference in the form of presentation of the disease produced by the different variants of CPV-2 [30]. The above added to the fact that there is no clarity about which are the variants that are circulating in the country and in what proportion are these [31].
The nucleotide sequencing confirms the above, since all the sequences obtained correspond to different strains of all the CPV-2 variants that are described in the BLAST™ database. Therefore, obtaining a positive PCR result, which would indicate the presence of any of the CPV-2 variants, is enough to determine the establishment of a support treatment and the application of sanitary measures that prevent the spread of the disease. virus. One of the disadvantages of using conventional PCR is the difficulty in differentiating whether the virus spread in the stool is due to the vaccine or a natural infection. Although it has been observed that a greater number of amplicons after a PCR could be indicative of a natural infection, this is not a reliable criterion due to the sensitivity of the molecular tests [32], so a result positive with this technique, should go in concomitance with the clinical signs of the patient.
New diagnostic methods have been studied to detect the presence of CPV-2 such as the quartz crystal microbalance [33] and the Loop-mediated Isothermal Amplification of nucleic acids [34], which They stand out for their simplicity and speed, but they are still in the standardization stage. Multiple real-time PCR techniques have also been developed that, in addition to detecting and quantifying the virus, also allow the identification of the variable present [29]. Additionally, the 12 nucleotide sequences obtained were aligned using the Bio Edit program version 7.2.6.1 [35] and entered the MEGA program version 7 [36] which allowed obtaining the percentage of variability between the CPV-2 fragments, which, on average, was 0.7%. This would indicate that this segment of the viral genome is highly conserved among the different variants of the virus and would explain the high percentage of nucleotide identity obtained with the different strains of the CPV-2 variants found in BLAST™.
Likewise, the primers used in this work were entered the BLAST™ program and it was found that, theoretically, they can detect FPV, generating a fragment like that produced with CPV-2, about 600bp. Consequently, the technique implemented in this work could also be applied to the diagnosis of FPV. This would be explained by the high similarity of the genome of the parvovirus grouped within the carnivorous Protoparvovirus species 1 [37], since the difference in the VP2 capsid protein of the FPV and the original CPV-2 does not they are more than six amino acids [10]. This should be confirmed with further studies. The management of a disease and the control measures that can be taken depend on the correct identification of the causative agent and according to the results obtained in this work, conventional PCR is effective in the detection of CVP-2 and although its realization has a higher cost in equipment, reagents and personnel, its sensitivity and specificity far exceeds the rapid tests used routinely in the veterinary clinic.
Therefore, while diagnostic methods that are currently under development do not become standardized and reliable methods, conventional PCR will continue to be an effective diagnostic method. It is also important to point out that the present study is the first molecular approach to CPV-2 that is carried out in the country. There are still studies to be done that allow us to contribute to the understanding of the evolution of CVP-2 and to determine which variants of the virus are circulating and their distribution in the country [38, 39].
Conclusion
The conventional PCR technique for the detection of CPV-2 was successfully implemented. Additionally, the superiority in terms of sensitivity of conventional PCR to IC that is commonly used in practice was demonstrated once again. Therefore, a negative HF result should not be considered as definitive and should be confirmed by a PCR test.
References
- Mylonakis ME, Kalli I, Rallis TS (2016) Canine parvoviral enteritis: an update on the clinical diagnosis, treatment, and prevention. Veterinary medicine, Auckland, New Zealand 7: 91-100.
- Berrios P (2001) Evolution and epidemiology of influenza virus, canine parvovirus type 2 and Nipah virus.
- Goodard A, Leisewitz A (2010) Canine Parvovirus. Vet Clin Small Anim 40: 1041-1053.
- Nandi S, Kumar M (2010) Canine Parvovirus: Current Perspective. Indian J Virol 21(1): 31-44.
- Sykes JE (2013) Canine Parvovirus Infections and Other Viral Enteritides. In: Canine and Feline Infectious Diseases. Elsevier St Louis, USA pp.141-151.
- Faz M, Martínez J, Gómez L, Quijano HI, Fajardo R, et al. (2019) Origin and genetic diversity of canine parvovirus 2c circulating in Mexico. Arch Virol 164(2): 371-379.
- Truyen U (2006) Evolution of canine parvovirus-a need for new vaccines? Vet Microbiol 115(1): 9-13.
- Hoezler K, Parrish C (2010) The emergence of parvoviruses of carnivores. Vet Res 41(6): 39.
- Bounavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, et al. (2001) Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. 82(12): 3021-3025.
- Decaro N, Bounavoglia C (2012) Canine parvovirus-A review of epidemiological and diagnostic aspects, with emphasis on type 2c. Ve Microbiol 155(1): 1-12.
- ICTV (2014) International Committee on Taxonomy of Viruses. Virus Taxonomy, Washington, DC, USA.
- Tinky S, Ambily R, Nair S, Mini M (2015) Utility of a rapid immunochromatographic strip test in detecting canine parvovirus infection compared with polymerase chain reaction. Vet World 8(4): 523-526.
- Parrish C (2011) Parvoviridae. In: Maclachlan J, Dubovi E (Eds.). Fenner's Veterinary Virology 4th (edn), Academic Press. San Diego, USA pp.225-235.
- Carter J, Saunders V (2007) Parvoviruses (and other ssDNA viruses). In: Virology: principles and applications. John Wiley & Sons West Sussex Inglaterra, UK pp.137-146.
- Humm K, Hughes D (2009) Canine Parvovirus Infection. In: Silverstein D, Hopper K (Eds.). Small Animal Critical Care Medicine. Saunders St Louis, USA pp.482-485
- Li R, Humm K (2015) Canine Parvovirus Infection. In: Silverstein D, Hopper K (Eds.). Small Animal Critical Care Medicine. 2da ed. Saunders St Louis, USA pp. 509-513.
- Cohn L, Langdon P (2008) Viral Infection. In: Morga, R (Ed.). Handbook of Small Animal Practice 5th (edn), Saunders. St Louis, USA pp. 1087-1103.
- Sherding R (2006) Intestinal Viruses. In: Bichard S, Sherding R (Eds). Saunders Manual of Small Animal Practice 3rd (edn), Saunders St Louis, USA pp.158-167.
- Pratelli A, Cavalli G, Normanno Mg, Palma G, Pastorelli A, et al. (2000) Immunization of pups with maternally derived antibodies to canine parvovirus (CPV) using a modified-live variant (CPV-2b). J Vet Med 47(4): 273-276.
- SAG (2015) Agricultural and Livestock Service. Veterinary medicines. Santiago Centro, Santiago, Chile.
- Desario E, Decaro N, Campolo M, Cavalli A, Cirone F, et al. (2005). Canine parvovirus infection: Which diagnostic test for virus? J Viral Method 126(1-2): 179-185.
- Cavalli A, Bozzo G, Decaro N, Tinelli A, Aliberti A, et al. (2001) Characterization of a canine parvovirus strain isolated from an adult dog. New Microbiol 24(3): 239-242.
- Markovich J, Stucker K, Carr A, Harbison C, Scarlett J, et al. (2012) Effects of canine parvovirus strain variations on diagnostic test results and clinical management of enteritis in dogs. J Am Vet Med Assoc 241(1): 66-72.
- Schmitz S, Coenen C, Koning M, Thiel H, Neiger R (2009) Comparison of three rapid commercial Canine parvovirus antigen detection tests with electron microscopy y polymerase chain reaction. J Vet Diagn Invest 21: 344-345.
- Mohyedini S, Jamshidi S, Rafati S, Nikbakht G, Malmasi A, et al. (2013) Comparison of immunochromatographic rapid test with molecular method in diagnosis of canine parvovirus. Iran J Vet Med 7(1): 57-61.
- (wd)-Test Virus Parvovirus Canino (C.P.V.).
- Kantere M, Athanasiou L, Spyrou V, Kyriakis C, Kontos V, et al. (2015) Diagnostic performance of a rapid in-clinic test for the detection of Canine Parvovirus under different storage conditions and vaccination status. J Virol Method 215-216: 52-55.
- Decaro N, Desario C, Beall M, Cavalli A, Campolo M, et al. (2010) Detection of canine parvovirus type 2c by a commercially available in-house rapid test. Vet J 184(3): 373-375.
- Chirinos AM (2015) Practical guide to real-time PCR.
- Kaur G, Chandra M, Dwivedi P, Narang D (2016) Multiplex real-time PCR for identification of canine parvovirus antigenic types. J Virol Method 233: (1-5).
- Wilson S, Illambas J, Siedek E, Stirling C, Thomas A, et al. (2014) Vaccination of dogs with canine parvovirus type 2b (CPV-2b) induces neutralising antibody responses to CPV-2a and CPV-2c. Vaccine 32(42): 5420-5424.
- Berrios P, Duran C (2005) Principales enfermedades virales de los caninos. Situación en Chile. Monogr Electron 2(2): 68-93.
- Meggiolaro M, Ly A, Rysnik SB, Silva C, Zhang J, et al. (2017) MT-PCR panel detection of canine parvovirus (CPV-2): Vaccine and wild-type CPV-2 can be difficult to differentiate in canine diagnostic fecal samples. Moll Cell Probe 33: 20-23.
- Kim Y, Lim S, Choi S, Cho I, Park E, et al. (2015) A novel assay for detecting canine parvovirus using a quartz crystal microbalance biosensor. J Virol Method 219: 37-27.
- Sun Y, Yen C, Tu C (2017) Immunocapture loop-mediated isothermal amplification assays for the detection of canine parvovirus. J Virol Method 249: 94-101.
- Hall T (1999) Bio Edit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95-98.
- Kumar S, Stecher G, Tamura K, (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870-1874.
- Shackelton L, Parrish C, Truyen U, Holmes E (2005) High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci 102(2): 379-384.
- BLAST (2017) Basic Local Alignmente Search Tool.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.