Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Nested Polymerase Chain Reaction for Identification of Mitochondrial Cytochrome b Gene of Gazella Dorcas
*Corresponding author: Reem Rabie Mohammed Salih, Department of Clinical Medicine, Faculty of Veterinary Medicine, University of Khartoum, Sudan.
Received: October 10, 2019; Published: January 21, 2020
DOI: 10.34297/AJBSR.2020.07.001105
Abstract
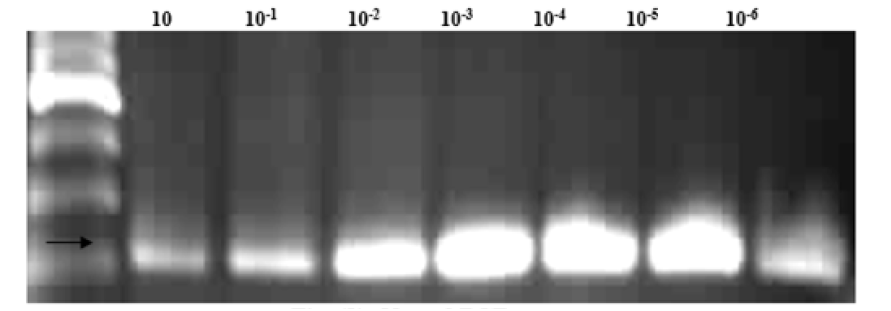
In this study nested PCR, was developed for rapid detection of mitochondrial cytochrome b gene. Whole blood in EDTA were collected from 17 Gazella dorcas from Hilat Kuku Zoo, Elkadaru’s farm and Mozamel Elkurdi’s farm in East of the Nile, Khartoum State. The origins of these animals are Dongola, River Nile- Northern Sudan and Butana- Central Sudan. The product was sent for sequencing to Macrogen Company- World Meridian 10F, Gasan-dong, Geumcheon-gu, Seoul, 153-781 Koreas. Nested PCR used DNA extracted from blood samples from Gazella dorcas by using GZ3 and GZ4 primers. The objective using this technique that nested- PCR is sensitive to detect the part of mitochondrial cytochrome- b gene of Gazella dorcas only. The primers were serially diluted (10 to 10-6) for sensitivity reaction. In all dilutions, the line was clear. The size of the product was 197 bp.
Keywords: Nested PCR; Blood; Dorcas; Gazell
Introduction
One of the most desert-adapted gazelles, dorcas gazelles may go their entire lives without drinking any water, obtaining all needed moisture from the plants which they eat [1]. They can withstand very high temperatures, although during hot weather they are primarily active at dawn, dusk, and throughout the night. Herds wander over large areas searching for food and tend to congregate in areas where recent rainfall has stimulated plant growth [1, 2, 3, 4 ].According to [5] dorcas gazelle still ranges in Algeria, Burkina Faso, Chad, Djibouti, Egypt, Eritrea, Ethiopia, Libya, Mali, Mauritania, Morocco, Niger, Somalia, Sudan, Tunisia and Western Sahara; its occurrence in Nigeria is very doubtful, while it is considered extinct in Senegal. Its geographical distribution was acquired from [6]. The polymerase chain reaction (PCR) is a scientific technique in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a DNA sequence. Developed [7, 8]. The method relies on thermal cycling, consisting of cycles of repeated heating and cooling of the reaction for DNA melting and enzymatic replication of the DNA. Primers containing sequences complementary to the target region along with a DNA polymerase are key components to enable selective and repeated amplification [8]. As PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the DNA template is exponentially amplified. PCR can be extensively modified to perform a wide array of genetic manipulations [8]. Nested polymerase chain reaction (Nested PCR) is a modification of polymerase chain reaction intended to reduce the contamination in products due to the amplification of unexpected primer binding sites. Polymerase Chain Reaction itself is the process used to amplify DNA samples, via a temperature-mediated DNA polymerase.
Anon (2010) [9] Nested PCR involves two sets of primers, used in two successive runs of (PCR). The second set intended to amplify a secondary target within the first run product [9]. A nested PCR has been developed by [10] to detect a 320 bp DNA segment of the gene encoding the p 57 protein. The sensitivity of the method was increased a hundredfold compared to a conventional PCR method. [10]. The rapidity, sensitivity and specificity of the nested PCR assay would greatly facilitate detection of mitochondrial cytochrome- b gene [11]. The nested PCR yielded a DNA band of expected 101 bp size. The NS1 gene primer sequences for partial length and nested PCR used in the study were the same as used earlier [8]. The nested PCR assay provides greater specificity because it involves 2 rounds of amplification. In the first round, a pair of external primers used to get a larger fragment, which acts as a template for second round of amplification with second set of internal primers [11]. This process provides an additional specificity to the reaction and greatly enhances the efficiency of amplification. The rapidity, sensitivity and specificity of the nested PCR assay would greatly facilitate detection of mitochondrial cytochrome- b gene [11].
Materials and Methods
Blood samples were collected from different species of animals (Gazelles, sheep, goats, cows, camels, donkeys, horses and pigs) in blood container with Ethylene diamine tetra acetic acid (EDTA) , these samples were used for DNA extraction. The extraction by used QiAgen commercial kit.
Extraction of DNA from Blood Samples:
For extraction of the DNA from blood samples we used commercial kit (QIAamp blood kit- QIAGEN Inc Chatsworth, CA, USA) according to the manufacturer´s instructions. Briefly, 200 μl from the Lysing buffer (L.pin sterile epindorf tube, then added 200 μl from the blood sample and added 10 μl from lyses enhancer, vortexed the tubes then incubated in water bath at 70ºC for 10 minutes. Add 200 μl absolute alcohol, vortexed and incubated in water bath at 70ºC for 2 minutes. Transferred all contents from the epindorf tubes and put to QIA spin column. Spin at 8000 rpm for 2 minutes and discarded the deposite. Washed with 500 μl buffer one, vortexed and spin at 8000 rpm for 2 minutes. Changed the QIAamp column and washed with 500 μl buffer two, vortexed and spin at 12000 rpm for 3 minutes. Discarded the deposit and put the QIAamp column in epindorf tube and add 200 μl from elution buffer and left for 1 minute. Put in the spin for 8000 rpm for 2 minutes then discard the column and saved the epindorf tube at freezing.
Nested Polymerase Chain Reaction
For detecting the mitochondrial cytochrome-b gene for dorcas gazelle we designed the primers by using bioedit software by accession number JN410257.1 of Gazella dorcas isolate from west 7 cytochrome- b (cytb) gene, complete cds; mitochondrial Tunisia from GenBank. The target DNA undergoes the first run of polymerase chain reaction with the first set of primers. The product from the first reaction undergoes a second run with the second set of primers, included bases of the positive sense strand ´5: CAC ACT CCT AGT TCT ATT CT. Complementary included bases of the complementary stand ´5: GTA TAA GGA TTA AGA TTA GA. Solved the primers: The left solved 371 μl, the total volume 100 pmol/ μl to final concentration volume 371 μl × 2 = 742 pmol/ μl. The complementary primer solved 289 μl, the total volume 100 pmol/ μl to final concentration volume 289 μl × 2 = 578 pmol/ μl. Fortytwo μl PCR mix were taken and mixed with 2 μl primers, 1 μl Taq DNA polymerase and 5 μl PCR product from first run (GZ1 and GZ2) for dorcas gazelle put in thermal-cycler tubes. All PCR amplification reaction was carried out in a final volume 50 μl.
Nested Polymerase Chain Reaction Sensitivity
To test the sensitivity of species-specific primer, serial dilution of the product from 10 μl to 10 -6 μl were subjected to the reaction containing the individual species DNA. Vortexed the mixture and adjusted the thermal-cycler apparatus at the specific program according to Tm. The thermal cycling profiles were as follows: at 95ºC, followed by 40 cycles of 94ºC for 1 min, 54ºC for 30 sec, and 72ºC for 45 sec and final incubation at 72ºC for 10 min. The total time of program was 2 hours and twelve minutes. After PCR took 10 μl from the product with 3 μl from loading dye mixed and loaded onto gel of 1.0% Seakem agarose (FMC Bioproduct, Rockland Me) and electrophoresed. Put the gel onto Ethidium bromide, the PCR products were easily identified under U.V light.
Discussion
Lack of detailed information about threatened groups of animals can hamper conservation efforts [12, 13]. Animal or meat species identification has been developed to address different concerns. Authentication of food ingredients is important for consumers because of food fraud was spread. The traceability of meat component in food improves consumer’s confidence in food products. The substitution of expensive meat with cheaper one is a major concern. For some consumer groups, such as vegetarians, the contamination of food with meat residue is strictly prohibited. Another good example of meat identification is the Halal food for the Muslim consumers, who are prohibited from consuming pork. In this study we used the tissues and cooked meat from Gazella dorcas to detect (mtcy-b gene) using PCR and this in accord with [14, 15, [16, 17, 18] findings.
In this study we developed nested-PCR method for very low DNA content products like blood for detection of mitochondrial cytochrome- b gene in Gazella dorcas in Sudan to identify authentic blood, especially from expensive dorcas gazelle’s products because the technique was more sensitive, easier, and faster than the conventional PCR. The result of nested PCR in this study is in agreement with [11, 19, 20] findings.
Results
The results of this study identified mtcyt-b gene in blood from 10 dorcas gazelles and show the sensitivity and specificity of the synthesized primers with target DNA from gazelle and other animals’ species [21, 22]. Nested PCR shows (Table 1)(Figure 1).
References
- Wilson DE, Reeder DM (1993) Mammal Species of the World (2nd Edition). Washington: Smithsonian Institution Press, USA.
- Happold DCD (1987) The Mammals of Nigeria, Oxford: Clarendon Press, UK.
- Walther FR (1990) Gazelles and related species. In Grzimek's Encyclopedia of Mammals, Edited by SP Parker, New York (USA) 5: 462-484.
- Gazella Dorcas (1998) In African Mammals Databank-A Databank for the Conservation and Management of the African Mammals 1 and 2. IEA (Institute of Applied Ecology).
- East R (1996) Antelope survey update. IUCN/SSC Antelope Specialist Group n°
- Yom Tov Y, Mendelssohn H, Groves CP (1995) Gazella dorcas. Mammalian Species 491: 1-6.
- Bartlett JMS, Stirling DA (2003) Short History of the Polymerase Chain Reaction. PCR Protocols 226: 3-6.
- Kary Mullis (1993) Nobel Lecture.
- Anon (2010) Scholarly articles on nested polymerase chain reactions.
- Chase DM, Pascho RJ (1998) Development of a nested polymerase chain reaction for amplification of the p57 gene of Renibacterium salmoninarum that provides a highly sensitive method for detection of the bacterium in salmonid kidney. Dis Aquat Org 34: 223-229.
- Sasimanas U, Piyachat M, Komsan A, Duangtip A, Kornsorn S, et al. (2011) Identification of species (meat and blood samples) using nested-PCR analysis of mitochondrial DNA. African Journal of Biotechnology 10(29): 5670-5676.
- Avise JC (1989) A Role for Molecular-Genetics in the Recognition and Conservation of Endangered Species. Trends in Ecology & Evolution 4(9): 279-281.
- Vogler AP, Desalle R (1994) Diagnosing Units of Conservation Management. Conservation Biology 8(2): 354-363.
- Saez R, Sanz Y, Toldra F (2004) PCR-based fingerprinting techniques for rapid detection of animal species in meat products. Meat Sci 66(3): 659-665.
- Aida AA, Che Man YB, Wong CM, Raha AR, Son R (2005) Analysis of raw meats and fats of pigs using polymerase chain reaction for Halal authentication. Meat science 69(1): 47-52.
- KesmenZ, Sahin F, Yetim H (2007) PCR assay for the identification of animal species in cooked sausages. Meat Sci 77(4) :649-653.
- Tanabe S, Miyauchi E, Muneshige A, Mio K, Sato C, et al. (2007) PCR method of detecting pork in foods for verifying allergen labeling and for identifying hidden pork ingredients in processed foods. Biol Sci Biotechnol Biochem 71(7): 1663-1667.
- Haunshi S, Basumatary R, Girish PS, Doley S, Bardoloi RK, et al. (2009) Identification of chicken, duck, pigeon and pig meat by species-specific markers of mitochondrial origin. Meat Sci 83(3): 454-459.
- Robert LH, William M, Benito F, Osborn BM, Tim W, et al. (2001) Phylogenetic reanalysis of the Saudi gazelle and its implications for conservation. Conservation Biology 15(4): 1123-1133.
- Lerp H Wronski, T Pfenniger, M Plath M (2011) Aphylogeoraphic framework for the conservation of Saharan and Arabian Dorcas gazelles (Artiodactyla: Bovidae). Org Divers Evol 11: 317-329.
- Johannes AL, Jacob BB, Frederik WJ (2001) On the origin of meat-DNA techniques for species identification in meat products. Veterinary Sciences Tomorrow p. 2.
- Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning, A laboratory handbook, Cold Spring Harbor Laboratory, New York Cold Spring Harbor.




 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.