Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
A Comparative Study on Global Genetic Diversity and Population Genetic Analysis of Orf Virus Isolates from Outbreaks and it’s Implications for the Vaccine Development
*Corresponding author: Saeed Nazeri, Research and Development Laboratory, Vira Vaccine Companys, Iran.
Received: March 11, 2020;Published: April 08, 2020
DOI: 10.34297/AJBSR.2020.08.001281
Abstract
Introduction: Orf virus envelope gene (B2L) encodes a highly immunogenic protein that exhibits lipase activity. Since Orf virus strains change genetically, it is essential to have adequate information on geospatial genetic diversity of global B2L to design an appropriate contagious ecthyma vaccine.
Materials and Methods: For this purpose, we have investigated twenty-four Iranian Orf virus B2L genes that were recovered from tow outbreaks of contagious ecthyma in sheep and goats from four provinces of Iran and compared with related available global sequences in GenBank.
Results: The nucleotide sequence analysis of Iranian B2L showed the occurrence of 71 SNPs. In total, 17 amino acid substitutions were detected with 2 unique mutations, resulting in 9 haplo types that 6 of them were not reported in any other geographic regions. Tests for departure from neutral evolution showed purifying selection. Phylogenetic analysis of the B2L gene suggests that the Iranian isolates were closely related to Turkey, India, China, Taiwan, Finland and Brazil isolates.
Conclusion: The present analysis has shown that genetic diversity in the global Orf virus exhibits low-level diversity, purifying selection and geographic variation. These results are of practical significance for the strategic development and deployment of control measures in particular for development of an appropriate contagious ecthyma vaccine.
Keywords: Contagious Ecthyma; Orf Virus; B2l Gene; Genetic Diversity; Population Genetic
Abbreviations: CPE: Cytopathic Effects; DMEM: Dulbecco’s Modified Eagle Medium; SNP: Single Nucleotide Polymorphism; Fst: Fixation Index; BPSV: The Bovine Papular Stomatitis Virus; PCPV: Pseudocowpox Virus; Nt: Nucleotide; AA: Amino Acid
Introduction
Contagious ecthyma (Orf, contagious pustular dermatitis, infectious labial dermatitis, scabby mouth, sore mouth) is a viral disease that is caused by the epitheliotropic Orf virus and it primarily causes disease in goats, sheep, and other ruminants globally [1-3]. The disease is also transmissible to humans (zoonotic) due to direct or indirect contact to people working with animals [4] and infections are manifested by painful lesions in hand and can spread to other organs [5]. The disease in sheep and goats are characterized by inflammatory, proliferative and scabby lesions in the lips, oral and nasal mucosa and muzzle. The lesions are painful and depending on the location of the lesions, animals may be unwilling to nurse, eat or walk [6]. Although, vaccination with attenuated Orf virus is the most common method to prevent and control the disease [7,8] there are recent reports revealing that the vaccination with the live attenuated virus cannot provide effective protection [9] suggesting that the Orf virus strains might have been changed genetically [10-12]. The virus is a member of Parapox virus genus which belongs to the Poxviridae family. It has a linear double-stranded DNA genome (134-139kb in size) [13]. The central region of the genome encodes proteins involved in the virus structure and assembly, while in the terminal regions of the genome there are genes associated with virulence, pathogenesis, host range [14,15]. The envelope gene (B2L) of the virus encodes a highly immunogenic major envelope protein of molecular weight about 42kDa and exhibits lipase activity [16]. This gene has been widely used for molecular characterization and phylogenetic analysis of isolates of Orf virus [8,17,18].
In recent years, outbreaks of Orf virus have occurred worldwide [12]. Although outbreaks of Orf virus have occurred in Iran and have been confirmed, there is no report available of the detailed sequence and phylogenetic analysis of the B2L gene of Orf virus in Iran in comparison with other isolates from other regions of the world. Having this data may resolve the determinants of gene flow and patterns of genetic diversity in global endemic regions. Therefore, in the current investigation, the main objective was to investigate the sequence diversity and population genetic of the B2L gene among natural Orf virus isolates. On the other hand, we extracted worldwide B2L sequence data available in the GenBank, and along with Iranian B2L sequence data, the global genetic diversity of this gene was evaluated. The obtained results would add more data on available global information regarding B2L genetic diversity and would also allow comparing the Orf virus population from this part of the world with those found in distinct and contrasting epidemiological settings to identify polymorphisms. The results of this study may be helpful for developing an appropriate live attenuated contagious ecthyma vaccine.
Material and Methods
Sampling of Orf Virus from Sheep and Goats
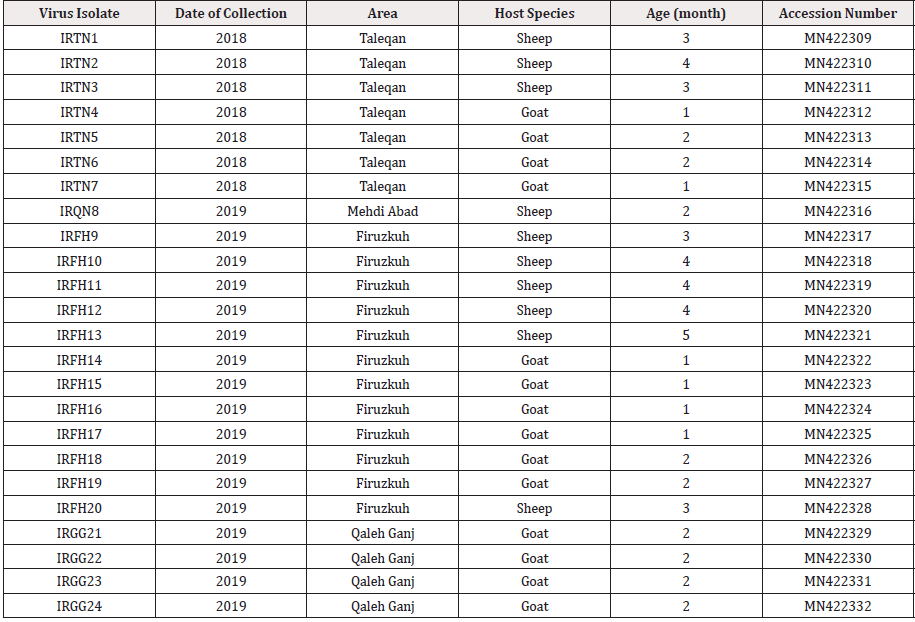
In the current investigation, 24 mouth scab samples were collected from two outbreaks of Orf virus which occurred in sheep and goats of Northwest (Taleqan in Alborz province; Mehdi Abad in Qazvin province; Firuzkuh in Tehran province) and Southeast (Qaleh Ganj, Kerman province) Iran during 2018-2019 (Figure 1, & Table 1). The outbreaks were characterized by the presence of cutaneous lesions on lips with no mortality. Sampling was performed using sterile forceps individually for each animal and the samples were immediately immersed into sterile PBS supplemented with 1% antibiotics (Penicillin 1000 IU-Streptomycin 10mg; Sigma, Germany). The samples were chilled on ice during the transport to the laboratory of Vira Vaccine Co. and were stored at -20°C for subsequent use.
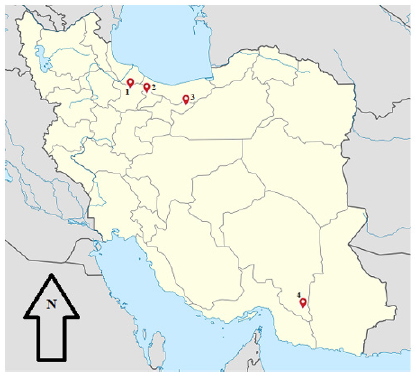
Figure 1: Geographic locations of the Orf virus infection in sheep and goats. Map of Iran showing the geographical sites where the 24 mouth scab samples have been collected during Orf suspected outbreaks from clinically sick sheep and goats in: a) Qazvin province (Mehdi Abad), b) Alborz province (Taleqan), c) Tehran province (Firuzkuh), d) Kerman province (Qaleh Ganj).
Virus Isolation
The collected mouth scab samples were washed three times with sterile PBS, cut into small pieces, ground, and suspended (1:10) in sterile PBS supplemented with antibiotics. The suspension was centrifuged at 5000 rpm for 10min and the supernatant was used to infect primary lamb testis (LT) cells. 200μL of the supernatant was inoculated onto a confluent layer of LT cells grown in a 25cm2 tissue culture flask containing 10ml Dulbecco’s Modified Eagle Medium (DMEM, Sig ma-Aldrich) supplemented with 10% fetal bovine serum (Gibco). The inoculated cultures were incubated in 37°C and 5% CO2 for 120h. The flasks observed daily for the presence of cytopathic effects (CPE). Samples were considered negative when no CPE was observed following three blind passages.
Viral DNA Extraction
The pathological tissue homogenates (10% w/v in PBS) were centrifuged at 10,000g for 10min at +4°C. The supernatants were used in order to viral DNA isolation. Virus genomic DNA was extracted using the commercially available DNA extraction kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer’s instruction. The purified DNA was dissolved in TE buffer and stored at -20°C until further testing.
Amplification and Sequencing of B2L Gene
For molecular analysis of B2L gene, PCR amplification was performed using a forward outer primer (B2LF: 5′- GACCTTCCGCGCTTTAATTT-3′) and a reverse primer (B2LR: 5′- CCCGCCTGCTAAAAGACT-3′) [19]. PCR amplification was performed in a final volume of 25μl reaction containing 125 μM each of dNTPs, 2 mM MgCl2, 0.2nM each of primers, 0.5 U Pfu DNA polymerase (Invitrogen, Carlsbad, CA, USA), and 1 μl DNA as template isolated from the tissue homogenates. The cycling condition for the amplification of the fragment was 95°C for 4 min followed by 30 cycles of 95°C for 30s annealing at 57°C for 45s, extension at 72°C for 45s, and final extension at 72°C for 7min. For each PCR run, a negative control consisting of 5μL of water instead of the template DNA was included. PCR fragment was analyzed by electrophoresis on 1% agarose gels (Invitrogen, Carlsbad, CA, USA).
In the present study, all 24 samples were subjected to sequencing analysis. The amplified products of B2L genes were purified using Qiagen DNA purification kit (Qiagen, Germany) according to the manufacturer’s instructions. Direct sequencing of the full-length of B2L was performed in both directions using B2LF and B2LR primers by the dideoxy chain termination procedure (Chemistry V3.1, Applied Biosystems) and the 3730XL DNA analyzer (Applied Biosystems) by Bioneer Sequencing Service (Korea). The obtained sequences were confirmed by Blast server https://blast.ncbi.nlm. nih.gov/Blast.cgi. https://blast.ncbi.nlm. nih.gov/Blast.cgi. All the sequences were analyzed and edited manually using BioEdit software version 7.2.5.
Sequence Analysis
The full-length B2L gene sequences of the Iranian Orf virus isolates were translated into amino acid sequences by Gene Runner software version 5. Then, the nucleotide and translated amino acid sequences were aligned with the corresponding NZ2 sequence (U06671) as a reference strain using the CLUSTALW method https://www.genome.jp/tools-bin/clustalw to determine the single nucleotide polymorphisms (SNPs). Moreover, the haplotypes were classified based on DNA and amino acid sequences of the examined samples in comparison with the NZ2 isolate. In addition, BLAST searches https://blast.ncbi.nlm.nih.gov/Blast.cgi were performed to compare these B2L haplotypes with other available sequences in the GenBank database. In this study, the nucleotide sequence data of B2L gene is available in GenBank databases under the accession numbers MN422309 to MN422332. DNA sequences from the Iranian B2L gene (Table 1) and DNA sequences from global B2L gene that previously deposited in the GenBank (Table 2) were prepared in FASTA format using CLUSTAL X version 2.0 and then analyzed in DnaSP version 5.10.01. The haplotype diversity (Hd), number of haplotypes (H), nucleotide diversity (π) and the minimum number of recombination events (Rm) were calculated using the DnaSP software.
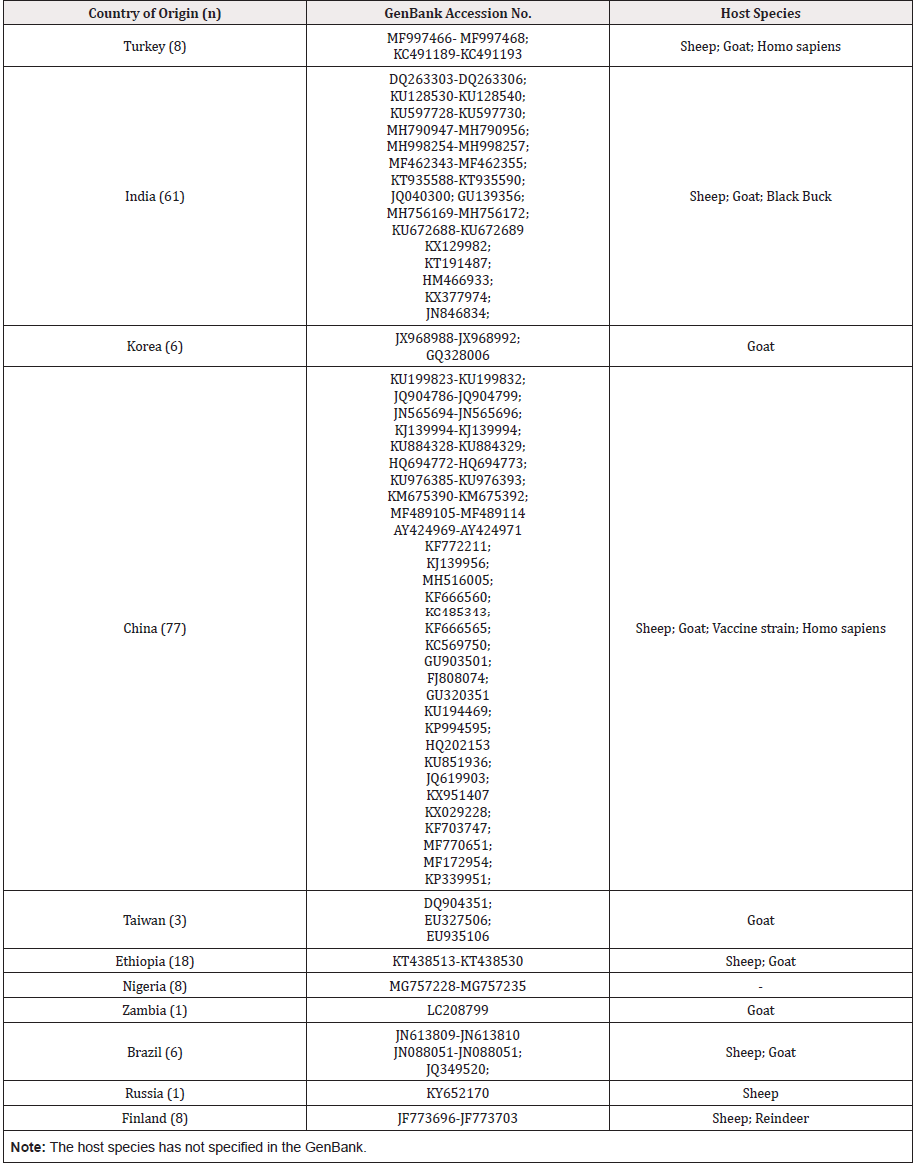
Table 2: Information about Orf virus isolates (n = 197) used for sequence and population genetic analysis.
To estimste the effect of natural selection on the generation of diversity in B2L within Iranian Orf virus isolates, the number of synonymous (dS) and nonsynonymous (dN) substitutions per site were performed using DnaSP version 5.10.01 based on the method of Nei and Gojobori’s (1986) [20] with the Jukes and Cantor (JC) correction (1969) [21]. Differences between dN and dS rates (dNdS) were evaluated by the Z-test using MEGA 6.0 software [22]. AP value less than 0.05 was considered statistically significant. To test the neutral theory of evolution based on intraspecific comparisons were done by Tajima’s et al. [23,24] D* and F* tests [24] using the DnaSP software. Tajima’s et al. [25] D test determines the departure from neutrality by comparing the estimations of nucleotide divsersity (θ) derived from the average pair wise diversity (π) and the total number of polymorphic sites (S) [25]. The Fu et al. [24] tests were also used to identify the departure from neutral patterns of nucleotide substitutions as deviations between the estimates of θ derived from the number of phylogenies compared to either the total number of mutations (D*) or the average pairwise diversity (F*) [24]. A positive value of Tajima’s et al. [24,25] D tests corresponds to positive natural selection, whereas a negative value signifies an excess of low-frequency polymorphisms relative to expectation, indicating population size expansion and/or negative/purifying selection. To examine whether deviation from neutrality occurs at certain residues of B2L, a sliding window analysis of Tajima’s D [25] across the coding sequences was done using a window size of 250bp and a step size of 10bp [26].
Population Genetic Analysis
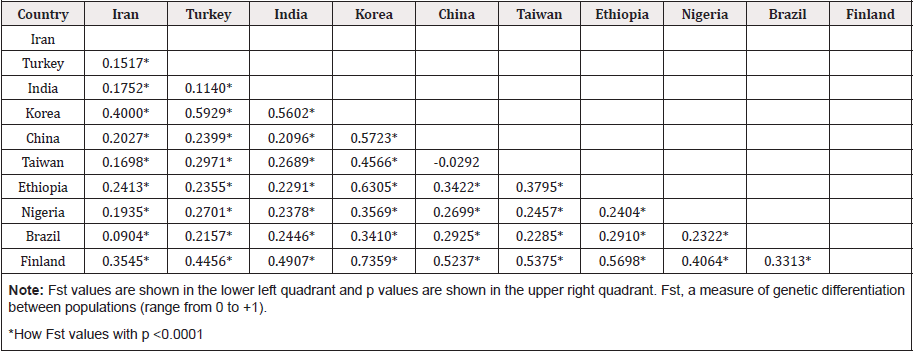
The genetic difference among the isolates of different endemic regions was measured by the fixation index (Fst) and calculated from the average number of pairwise DNA sequence diversity interspecific and intraspecific populations using DnaSP software. Fst analysis was performed based on B2L sequences published in GenBank (nt: 1-1137), including Turkey (n=8), India (n=61), Korea (n=6), China (n=77), Taiwan (n=3), Ethiopia (n=18), Nigeria (n = 8) Brazil (n=6) and Finland (n=8). The Fst measures the proportion of overall allele diversity attributable to differences within populations, and its interpretation at each locus was based on four categories defined earlier as no differentiation (0), low genetic differentiation (0-0.05), moderate differentiation (0.05-0.15) and high differentiation (0.15-0.25) [27]. P≤0.05 was considered as significant difference. Finally, phylogenetic analysis was generated using the neighbor-joining method with Kimura 2-parameter distance [28] in MEGA 6.0 software. The sequences generated in this Study (24 sequences) as well as Turkey (8 sequences), India (61 sequences), Korea (6 sequences), China (77 sequences), Taiwan (3 sequences), Ethiopia (18 sequences), Nigeria (8 sequences) Brazil (6 sequences), Finland (8 sequences), Russia (1 sequence) and Zambia (1 sequence), were analyzed along with reference sequences (BPSV; the bovine papular stomatitis virus and PCPV; pseudocowpox virus) retrieved from GenBank.
Results
Identification of Orf Virus
To confirm the Orf virus identity, total DNA was extracted from the mouth scab samples and screened by PCR amplification. A PCR product of 1,210 bp (open reading frame of 1,137 bp) was visualized on 1% agarose gel (Figure 2) from all samples. This result confirmed the presence of infectious Orf virus in all attended herds. Moreover, after inoculation of LT cells with the supernatants from the clinical samples, characteristic poxvirus-induced CPE (with rounded cell detaching from monolayer) developed after 2-4 days in culture for either the first or second blind passage (Figure 3A). In the negative control no CPE was observed (Figure 3B).
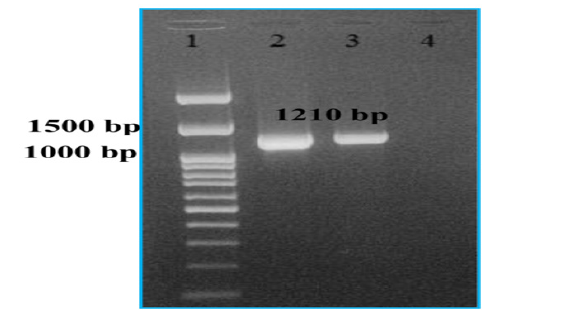
Figure 2: PCR amplification of B2L gene of sheep Orf virus isolate IRTN1 and goat Orf virus isolate IRTN4. The complete coding regions of IRTN1 isolate (lane 2) and IRTN4 isolate (lane 3) amplified from viral genomic DNA. Lane 4: negative control. Lane 1 (left): 100 bp-DNA ladder plus (cinnagen).

Figure 3: Light microphotographs of LT cell cultures and Orf virus. a) Cytopathic effects of LT cells after Orf virus infection; magnification: 100 × b) Not infected LT cells; magnification: 100 ×
Sequence analysis of Iranian and global B2L gene sequences
In the current study, the complete B2L gene (24 samples) were amplified, sequenced and used for comparative analysis. The nucleotide sequence analysis of Iranian B2L gene sequences in comparison with the NZ2 sequence (the GenBank accession number U06671) showed the occurrence of 71 SNPs (including 54 synonymous and 17 non-synonymous) (Table 3) and the results of multiple alignments of B2L nucleotide (nt) and amino acid (aa) sequences demonstrated that SNPs were randomly distributed (Table 3). Based on the full-length comparison of amino acid (aa: 1-378) alignment of all 24 Iranian isolates with the NZ2 B2L amino acid sequence, 9 distinct haplotypes were detected (Table 3). Interestingly, haplotypes were differed between endemic areas for example haplotypes IRB2L1 and IRB2L2 were exclusively found in Alborz province, haplotype IRB2L3 in Qazvin province, IRB2L4-IRB2L8 haplotypes in Tehran province and IRB2L9 in Kerman province (Table 3). Moreover, using BLAST search in the GenBank database, the comparison of the amino acid sequences of the isolates in the present study with previously reported B2L haplotypes showed that all of the haplotypes excluding IRB2L1, IRB2L2 and IRB2L3 were new and not reported before. The results showed a high degree of sequence identity at the amino acid level (100%) between Orf virus isolates from Alborz province (IRTN1, IRTN3, IRTN5, IRTN7 isolates) with an Indian isolate (GenBank accession No. AMY16340), Orf virus isolates from Alborz province (IRTN2, IRTN4, IRTN6) with a Turkish isolate (GenBank accession No. AGG86677) and Orf virus isolate from Qazvin province (IRQN8 isolate) with a Chinese isolate (GenBank accession No. ADF28645). Based on the full sequence of B2L protein (aa:1-378) reported in the GenBank (221 sequences previously reported), 186 amino acid replacements have been already identified in global Orf virus isolates from different endemic regions, in which 2 of them (S32N, W158C) were new and they have been detected in the present study (Table 3). Unique point mutation S32N was observed in haplotypes IRB2L6 and IRB2L7 (Tehran isolates) and W158C was observed in haplotype IRB2L9 (Kerman isolates).
The haplotype and nucleotide diversity for the B2L gene within the studied isolates were 0.924 ± 0.026 and 0.01898±0.00158, respectively. The isolates from Nigeria revealed the highest haplotype and nucleotide diversity per site (1.000±0.063 and 0.0353 ±0.00501, respectively) (Table 4). Moreover, the lowest haplotype and nucleotide diversity per site were shown in isolates from Taiwan (0.667 ± 0.314) and Korea (0.00475 ± 0.00106), respectively (Table 4). Estimation of the parameter Rm in the B2L locus reveals 27,25, 9,7,3 and 3 recombination sites in virus populations from India, China, Iran, Nigeria, Turkey and Brazil, respectively whereas Korea, Taiwan, Ethiopia and Finland isolates had no potential recombination sites (Rm=0) (Table 4). These results suggest that recombination could shape diversity at the B2L locus.
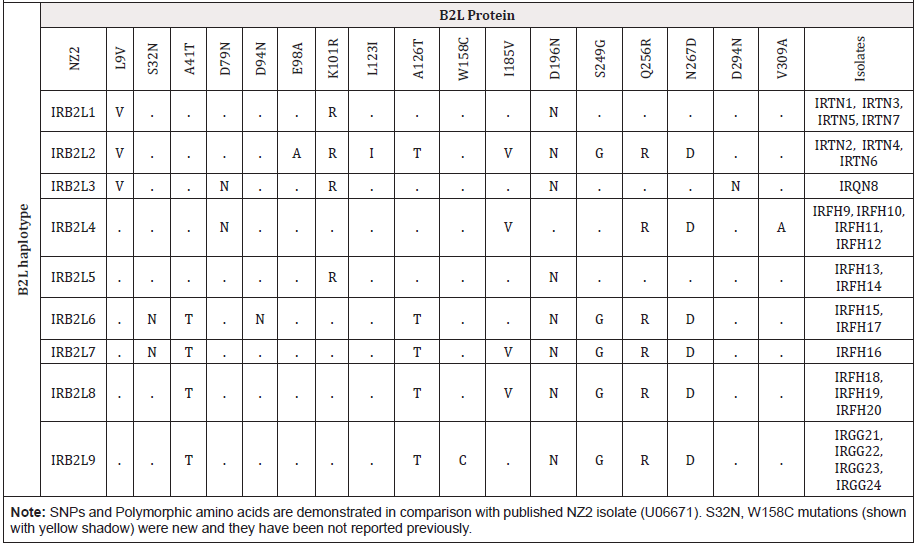
Table 3: Single nucleotide polymorphisms and amino acid substitutions of B2L in Iranian Orf virus isolates comparing to NZ2 as reference sequence (accession no. U06671).
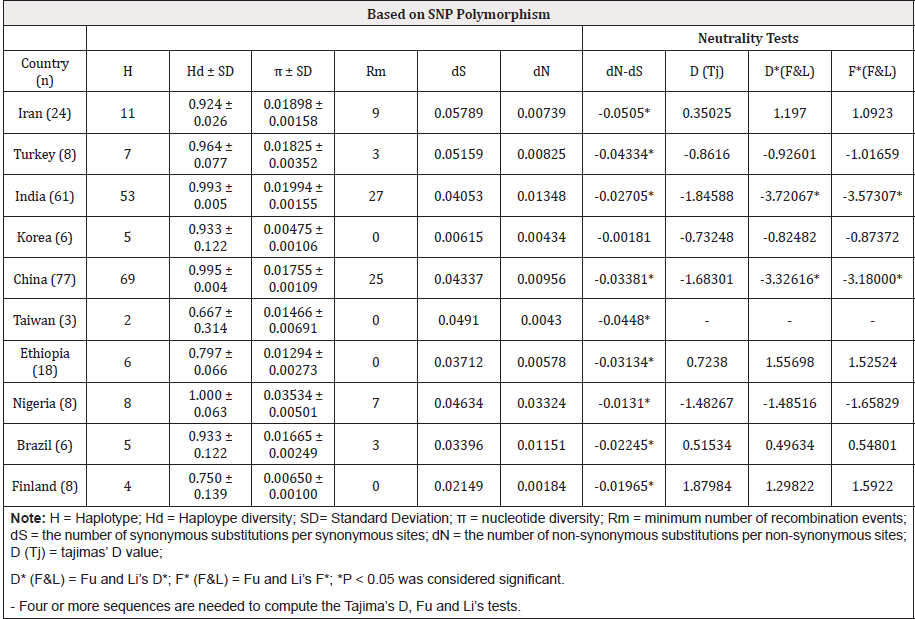
Table 4: DNA sequence polymorphism analysis for global B2L gene and tests for departure from neutrality.
Tests for Departure from Neutral Evolution
Tests for departure from neutral evolution analysis of patterns of nucleotide substitutions in the B2L sequences of isolates in this study reveals that dS significantly exceeds that dN in all virus populations (Z test, P<0.05) except Taiwan (Z test, P>0.05) (Table 4). Meanwhile, statistics based on Tajima’s D [25] did not show evidences of departure from neutral expectation (Table 4). However, sliding window analysis of Tajima’s D [25] across the coding sequences revealed significant negative at certain residues in the B2L gene in Orf virus populations from India, China, Finland and Nigeria, suggesting purifying selection in the B2L gene (data not shown). Moreover, statistics based on Fu et al. [24] D* and F* did not show evidences of departure from neutral expectation except India (-3.72067 and -3.57307, respectively; P<0.05) and China (-3.32616 and -3.18000, respectively; P<0.05), suggesting purifying selection (Table 4).
Genetic Differentiation Between Global Orf Virus and Phylogenetic Tree Construction
We observed population differentiation between isolates from Alborz, Tehran and Kerman provinces in which the FST values revealed significantly high differentiation (Fst >0.15, P<0.0001), implying that they did not share a common origin. Moreover, Orf virus populations from different populations having full length sequences (1-1137) in GenBank were significantly different from one another, indicating strong differentiation and reproductive isolation among populations excluding Brazilian with Iranian Orf virus isolates (Fst = 0.0904, P < 0.0001 (Table 5) and Indian with Turkish Orf virus isolates (Fst = 0.1155; P<0.0001 (Table 5) that showed moderate differentiation. The phylogenetic tree constructed based on the full length of B2L gene revealed that all Iranian Orf virus isolates were clustered with Indian, Turkish, Korean, Chines, Taiwanese, Brazilian and Finnish isolates (Figure 4). Interestingly, the only Russian B2L sequence was clustered in the closely related branches with several Chinese, a Nigerian and a Brazilian isolates. Moreover, the Zambian B2L sequence was clustered in the closely related branches with several Korean, Iranian and a Taiwanese isolates. Most of Ethiopian and Nigerian Orf virus isolates seemed to be isolated and located in separate branches. The tree also revealed that several available B2L sequences from Turkey and china were clustered with Indian isolates. However, there was no clear lineage for each population because several isolates from different endemic areas were shared or placed in the closely related branches (Figure 4).
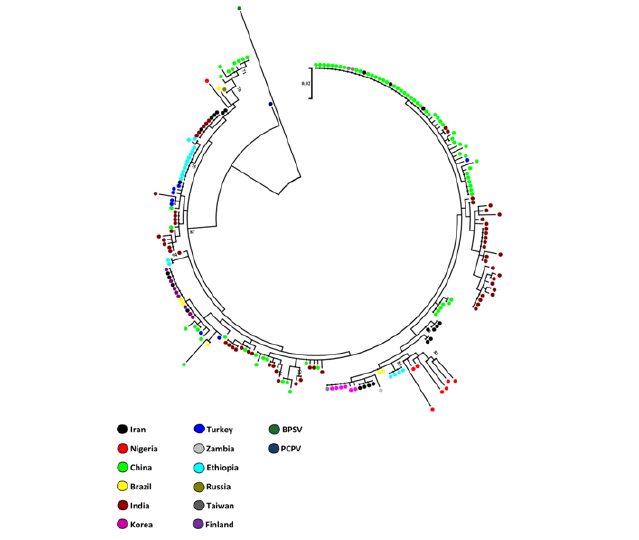
Figure 4: Phylogenetic tree based on nucleotide sequences of the B2L gene. A neighbor-joining tree was constructed by the MEGA 6 program based on Iranian B2L (n= 24) and the other sequences from endemic regions including; Turkey (n = 8), India (n = 61), Korea (n = 6), China (n = 77), Taiwan (n = 3), Ethiopia (n = 18), Nigeria (n = 8), Brazil (n = 6), Finland (n = 8), Zambia (n = 1) and Russia (n = 1). The bovine papular stomatitis virus (BPSV) and pseudocowpox virus (PCPV) applies as reference strains. The distance matrix was prepared using the Kimura 2-parameter evolutionary model. Alignment substitutions were considered for analysis, while gaps were excluded. The bootstrap values on the branches have been shown by the number of times in 1000 replications and bootstrap values above 70% are shown. The scale bar represents a genetic distance. Dots with different colors represent isolates and their geographic origins as listed on the lower left.
Discussion
Orf virus infection of sheep and goats is universally neglected globally because of the low morbidity rates. Although the disease is relatively benign, the outbreaks generate economic losses associated with a growth delay and deterioration of the animal’s body condition. In Iran, contagious ecthyma disease is endemic, and it has been reported in both sheep and goats. However, despite the increased frequency of the disease outbreaks, there hasn’t been any attempt to further inquire the disease. Sequence analysis of B2L has been used to address the genetic diversity and genetic relationships among Orf virus isolates to identify parapox virus species involved in clinical cases or outbreaks [29,30]. Moreover, in developing an efficient live attenuated contagious ecthyma vaccine, global information on circulating virus is necessary. In this regards, the current study describes for the first time, the genetic characterization of Orf virus isolates involved in two outbreaks in sheep and goats in Iran.
In this study, several nucleotide changes were synonymous whereas other resulted in change to an amino acid of a similar group (such as leucine to valine) or different group (such as tryptophan to cysteine) (Table 4). Whether these alterations result in differences in protein function is unknown. Overall the Iranian B2L gene sequences showed high similarity to previously sequenced foreign B2L gene. However, some of them had specific mutations (Table 3) that differentiated them from previously sequenced isolates [9,12,31,32]. The results showed a high difference of haplotype and nucleotide diversity exists among Orf virus isolates globally ranged from 0.667-1.000 and 0.00475-0.03534, respectively (Table 4) probably due to the different pattern of genetic variation and evolutionary relationships. However, in consistent with the previous report [33] a little variation observed within Orf virus isolates of countries individually. Gene evolution and genetic diversity in the B2L sequences could have been arisen from selective pressures and recombination between viruses. In this regards, the relatively higher levels of haplotype and nucleotide diversity of B2L from Iran, India, China, Nigeria, Turkey and Brazil than that from Korea, Taiwan, Ethiopia and Finland seems to be in line with recombination in each population (Table 4) and it is likely that recombination could further generate novel alleles. Furthermore, Tajima’s D [25] sliding window analysis revealed significant negative values at certain residues in the B2L gene in Orf virus populations from India, China, Finland and Nigeria, suggesting purifying selection at certain residues in the B2L gene. Moreover, in consistent with the only previous report [33] a significant difference between dN−dS indicated that the B2L is globally under purifying selection (Table 4). The results suggest that these populations may be undergoing a period of evolutionary expansion. Despite low level of sequence diversity, genetic distances inferred from the B2L locus among Orf virus populations could be deployed for analysis of population structure. In the present work, genetic differentiation of Orf virus population between Alborz, Tehran and Kerman Provinces was significant. Moreover, none of the detected haplotypes in the endemic regions of Iran were common. Therefore, diversity in the B2L locus of Orf virus in Iran seems to exhibit spatial variation.
Nevertheless, deeper molecular and serological analysis with more genome information is necessary to confirm this hypothesis and to better understand the circulation of the Orf virus isolates between domestic animals. Moreover, it is striking that in the regions where the isolates come from sheep and goats there were several different haplotypes (IRB2L1, IRB2L2 for Taleqan and IRB2L4-8 for Firuzkuh), while in the region where the isolates come from goat only there was a single haplotype (IRB2L9 for Qaleh Ganj). This finding is probably exhibits that in regions with a mixed population of sheep and goats, there is a greater presence of haplotypes. Interestingly, the worldwide genetic differentiation among Orf virus populations at B2L locus were significantly high (Table 5).
In this regards, gene flow between Orf virus populations may not occur because geographic distance has precluded intraand intercontinental spread of virus. Although Orf virus can be transferred between remote endemic areas through livestock movement, this situation could be inefficient due to rarity in numbers of such cases to contribute to genetic admixture of viruses. In the current study, phylogenetic analysis showed that Iranian B2L genes were more related to India, Turkey, Korea, China, Taiwan, Brazil and Finland Orf virus isolates (Figure 4) and the comparative sequence analysis revealed that the Iranian Orf virus isolates displayed more than 99% of identity at nucleotide level with the mentioned isolates. These findings would suggest a potential introduction of this Iranian Orf virus isolate from those countries, possibly through their economic activities in Iran. However, no concrete evidence to support this hypothesis is available. Moreover, Phylogenetic analysis has placed all Turkish isolates to several cluster and more related to northwest Iranian isolates, albeit without significant bootstrap support (Figure 4) suggesting possible trans-boundary transmission of the virus between the two countries. However, it seems that genetic diversity of the B2L locus displays more favor towards geographic variation.
Conclusion
The present analysis has shown for the first time, a genetic diversity and population genetic analysis of Orf virus utilizing the B2L gene in Iranian isolates and bring an insight into a knowledge of Orf virus is circulating throughout the world. The global B2L gene exhibits low level diversity, purifying selection and geographic variation that could affect the distribution of existing virus isolates. Negative selection and recombination have shaped diversity at the B2L locus. These results are used for understanding the nature of the Orf virus population in Iran and also are of practical significance for the strategic development and deployment of control measures in particular for development of an appropriate contagious ecthyma vaccine.
Acknowledgements
This study received funding from Vira Vaccine Shaya Company.
Conflict of Interest
All authors declare that they have no conflict of interest with respect to the research, authorship, and/or publication of this article.
References
- Guo J, Rasmussen J, Wünschmann A, De La Concha-Bermejillo A (2004) Genetic characterization of Orf viruses isolated from various ruminant species of a zoo. Vet Microbiol 99(2): 81-92.
- Vikøren T, Lillehaug A, Åkerstedt J, Bretten T, Haugum M, et al. (2008) A severe outbreak of contagious ecthyma (Orf) in a free-ranging musk ox (Ovibos moschatus) population in Norway. Vet Microbiol 127(1-2): 10-20.
- Inoshima Y, Ito M, Ishiguro N (2010) Spatial and temporal genetic homogeneity of Orf viruses infecting Japanese serows (Capricornis crispus). J Vet Med Sci 72(6): 701-707.
- Bayindir Y, Bayraktar M, Karadag N, Ozcan H, Kayabas U, et al. (2011) Investigation and analysis of a human Orf outbreak among people living on the same farm. New Microbial 34(1): 37-43.
- Duchateau NC, Aerts O, Lambert J (2014) Autoinoculation with Orf virus (ecthyma contagiosum). Int J Dermatol53(1): 60-62.
- Gumbrell RC, McGregor DA (1997) Outbreak of severe fatal Orf in lambs. Vet Rec 141(6): 150-151.
- Zhao K, He W, Gao W, Lu H, Han T, et al. (2011) Orf virus DNA vaccines expressing OrfV 011 and OrfV 059 chimeric protein enhances immunogenicity. Virol J 8: 562.
- Zhang K, Lu Z, Shang Y, Zheng H, Jin Y, et al. (2010) Diagnosis and phylogenetic analysis of Orf virus from goats in China: a case report. Virol J 7:78.
- Bora DP, Barman NN, Das SK, Bhanuprakash V, Yogisharadhya R, et al. (2012) Identification and phylogenetic analysis of Orf viruses isolated from outbreaks in goats of Assam, a northeastern state of India. Virus Genes 45(1): 98-104.
- Chan KW, Yang CH, Lin JW, Wang HC, Lin FY, et al. (2009) Phylogenetic analysis of parapoxviruses and the C-terminal heterogeneity of viral ATPase proteins. Gene 432(1): 44-53.
- Cottone R, Büttner M, Bauer B, Henkel M, Hettich E, et al. (1998) Analysis of genomic rearrangement and subsequent gene deletion of the attenuated Orf virus strain D1701. Virus Res 56(1): 53-67.
- Hosamani M, Bhanuprakash V, Scagliarini A, Singh RK (2006) Comparative sequence analysis of major envelope protein gene (B2L) of Indian Orf viruses isolated from sheep and goats. Vet Microbiol 116(4): 317-324.
- Chan KW, Lin JW, Lee SH, Liao CJ, Tsai MC, et al. (2007) Identification and phylogenetic analysis of Orf virus from goats in Taiwan. Virus genes 35(3): 705-712.
- Upton C, Slack S, Hunter AL, Ehlers A, Roper RL (2003) Poxvirus orthologous clusters: toward defining the minimum essential poxvirusJ virol 77(13): 7590-7600.
- Lateef Z, Baird MA, Wise LM, Young S, Mercer AA, et al. (2010) The chemokine‐binding protein encoded by the poxvirus Orf virus inhibits recruitment of dendritic cells to sites of skin inflammation and migration to peripheral lymph nodes. Cell Microbiol 12(5): 665-676.
- Sullivan JT, Mercer AA, Fleming SB, Robinson AJ (1994) Identification and characterization of an Orf virus homologue of the vaccinia virus gene encoding the major envelope antigen p37K. Virology 202(2): 968-973.
- Inoshima Y, Morooka A, Sentsui H (2000) Detection and diagnosis of parapoxvirus by the polymerase chain reaction. J Virol Methods 84(2): 201-208.
- Oem JK, Roh IS, Lee KH, Lee KK, Kim HR, et al. (2009) Phylogenetic analysis and characterization of Korean Orf virus from dairy goats: case report. Virol J 16(6): 167.
- Mayenga C (2015) Molecular diagnosis and characterisation of Orf virus in symptomatic goats in coast and Dar es salaam regions, Tanzania. (Doctoral dissertation, Sokoine University of Agriculture),
- Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3(5): 418-26.
- Jukes TH, Cantor CR (1969) Evolution of protein molecules. Mammalian protein metabolism 3:132.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12): 2725-2729.
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3): 585-595.
- Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133(3): 693-709.
- Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105(2): 437-460.
- Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11): 1451-1452.
- Balloux F, Lugon‐Moulin N (2002) The estimation of population differentiation with microsatellite markers. Mol Ecol 11(2): 155-165.
- Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2): 111-120.
- Cargnelutti JF, Flores MM, Teixeira FR, Weiblen R, Flores EF (2012) An outbreak of pseudocowpox in fattening calves in southern Brazil. J Vet Diagn Invest 24(2): 437-441.
- Sant’Ana FJ, Rabelo RE, Vulcani VA, Cargnelutti JF, Flores EF (2012) Bovine papular stomatitis affecting dairy cows and milkers in midwestern Brazil. J Vet Diagn Invest 24(2): 442-445.
- Zhang K, Liu Y, Kong H, Shang Y, Liu X (2014) Comparison and phylogenetic analysis based on the B2L gene of Orf virus from goats and sheep in China during 2009-2011. Arch Virol 159(6): 1475-1479.
- Venkatesan G, De A, Arya S, Kumar A, Muthuchelvan D, et al. (2018) Molecular evidence and phylogenetic analysis of Orf virus isolates from outbreaks in Tripura state of North-East India. Virus disease 29(2): 216-220.
- Sahu BP, Majeea P, Sahoob A, Nayaka D (2019) Molecular characterization, comparative and evolutionary analysis of the recent Orf outbreaks among goats in the Eastern part of India (Odisha). Agri gene 12:1



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.