Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Age-Related Features of Detectability of Xenobiotic Enzyme Gene Polymorphism in Pregnant Women with Fetal Growth Restriction Syndrome
*Corresponding author: Mavlyanova NN, Republican Specialized Scientific and Practical Medical Center of Obstetrics and Gynecology, Ministry of Health, Republic of Uzbekistan.
Received: March 18, 2020; Published: April 30, 2020
DOI: 10.34297/AJBSR.2020.08.001314
Keywords
Keywords: Bone fracture; Bone drilling and equipment Specialist; Knowledge transfer; Oil well drilling
Introduction
Fetal growth restriction syndrome (FGRS) is a clinical syndrome caused by morphological and functional changes in the placenta and penetrant by limit of the growth and development of the fetus, its hypoxia, that arise as a result of the combined reaction of the fetus and placenta to various disorders of the pregnant woman. This syndrome is based on pathological changes in the fetal and or uterin-placental complexes with a derangement of the compensatory-adaptive mechanisms at the molecular, cellular and tissue levels. In this case, the transport, trophic, endocrine, metabolic, antitoxic functions of the placenta underlying the pathology of the fetus and newborn are disordered [1-7]. The most significant risk factors of FGRS development include preeclampsia and a combination of pregnancy with extragenital pathology, accompanied by vascular damage. Various etiological factors, affecting at different stages of the development and functioning of the placenta, are ultimately involved in the general pathogenetic mechanism leading to the development of the fetal growth limit syndrome, one of the main manifestations of which is considered a violation of placental circulation - the main function of the placenta [8-16]. In this regard, the main direction in the study of the problem of FGLS, is the development of objective methods for prediction, preclinical diagnosis, optimal prevention and treatment, which can significantly reduce the frequency of FGLS, and its complications. Recently, special attention has been paid to studying the genes of xenobiotic biotransformation enzymes (XBEs), which are candidate genes for the formation of a predisposition to these pathologies, since their protein products interact with the environment, detoxifying or toxicizing foreign chemical compounds that enter the body, including also drugs [17-19]. However, they have not yet been sufficiently studied as genetic predisposition factors in fetal growth limit syndrome. According to the literature, these genes are a rather complex object of study due to a number of their specific features [11,14,20]. These are overlapping substrate specificity, inducibility and participation in the metabolism of endogenous compounds. But it is precisely these features of XBE that make it possible to assume that they can be genetic markers at all stages of the development of the disease from its initiation to the outcome and, accordingly, will make it possible to identify a predisposition, help in the early diagnosis of the disease, knowing the patient’s genotype, make a prognosis of the course of the disease, and choose the most suitable therapy.
Aim
The aim of our research was to study the detectability of allelic variants of gene polymorphism of xenobiotic biotransformation enzymes of pregnant women with fetal growth limit syndrome taking into account the age of the pregnant.
Materials and Research Methods
The object and subject of the study were pregnant women with fetal loss syndrome (FLS), DNA samples from patients and healthy donors, glutathione transferase genes GSTM1 (1p13.3), GSTT1 (22q11.2) and glutathione transferase gene GSTP1 (IIe 105 Val). The study included 143 pregnant women aged 19 to 34 years, observed at the clinic of Republican Specialized Scientific and Practical Medical Center for Obstetrics and Gynecology, Ministry of Health of the Republic of Uzbekistan (RSSPMCOG MH RUz). All pregnant women underwent general clinical, laboratory and functional studies according to the standard for diagnostics and therapy (2015). Molecular genetic testing of biomaterials (DNA) was carried out on the basis of the Department of Molecular Medicine and Cellular Technology Research Institute of Hematology and Blood Transfusion under the Ministry of Health of the Republic of Uzbekistan. The object and subject of the study were DNA samples of pregnant and healthy donors, glutathione transferase genes of the first phase - GSTM1 (1p13.3), GSTT1 (22q11.2) and the second phase - GSTP1 (IIe 105 Val). During genetic studies, the population control was used as a comparison group, which was represented by DNA samples (n=72) of conditionally healthy ones from the DNA bank of this department. DNA samples were isolated from peripheral blood lymphocytes in accordance with a modified methodology. The concentration and purity of the extracted DNA was evaluated by measuring the optical density of DNA-containing solutions at a wavelength of 260 and 280 nm against TE on a NanoDrop 2000 spectrophotometer (USA). Genotyping of GSTT1 and GSTM1 polymorphism was carried out by PCR on programmable thermal cyclers CG-1-96 Corbett Research (Australia) and 2720 Applied Biosystems (USA), using test systems of LLC Litekh (Russia), according to the manufacturer’s instructions.
The Results of the Study
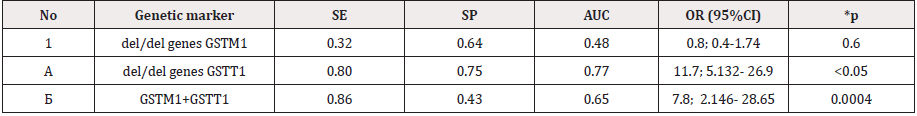
Clinical, laboratory and functional studies have shown that among the 143 pregnant women examined, fetal growth limit syndrome was detected in 105, which amounted to 73.4%. According to the severity degree, I - the degree of severity was diagnosed in 35 (33.3%), II the degree in 48 (45.7%) and III degree in 22, which amounted to 20.9%, respectively. Information on gene sequences and primer structure was obtained taking into account the original literary source [1] and Gene Bank. The characteristics of the genetic marker and the sequence of synthesized oligoprimers are shown in Table 1. Molecular genetic studies of the glutathione transferase genes GSTM1 (1p13.3), GSTT1 (22q11.2) and IIe 105 Val of the GSTP1 xenobiotic enzyme gene in the blood of pregnant women with FGLS revealed the following features of the distribution of alleles (Table 2) and genotypes of GSTM1 and GSTP1 gene polymorphisms (Tables 3 & 4). As it can be seen from Table 3, in the main group of pregnant women with SPP, functional alleles of GSTM1 “+” were detected in 67.8% of cases (40), while deletion variants (non-functional) of GSTM1 (0/0) were detected in 32.2% (19) cases. Whereas, functional allelic variants of GSTT1 “+” genotypes were detected in 20.3% of cases, and deletion variants in 79.6% (47) cases, respectively.
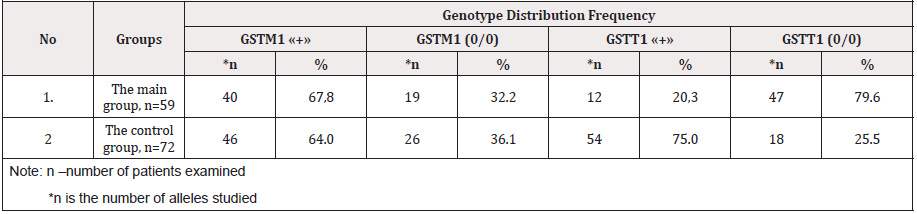
Table 3: Distribution frequency of alleles and genotypes of del / del genes polymorphism of the GSTM1 and GSTT1 in pregnant and control groups.
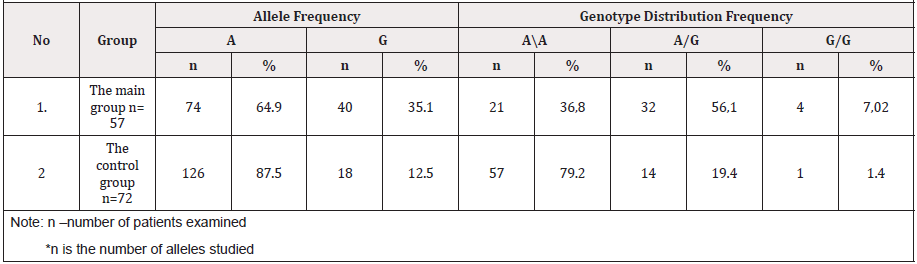
Table 4: The distribution frequency of alleles and genotypes of polymorphism IIe 105 Val of the GSTP1 gene in groups of patients and control.
As the comparative analysis of the distribution frequencies of the alleles and genotypes of the IIe 105 Val polymorphism of the GSTP1 xenobiotic enzyme gene among 114 DNA samples in 57 pregnant women revealed the presence of the normal A allele and 64.1% of the G allele in 35.1% of cases. Whereas, in the control group, the frequency of occurrence of the mutant allele IIe 105 Val of the GSTP1 xenobiotic enzyme gene was 12.5%, which was 2.8 times lower in comparison to the main group (P <0.05). For a detailed assessment of the prognostic criterion for the significance of the polymorphism of the genotypes of xenobiotic enzymes GSTM1, GSTT1 and GSTP1 in the development of fetal loss syndrome in pregnant women, we analyzed the results of analyzes depending on the presence of fetal loss syndrome (FGLS) and without .
The results of the study showed, that pregnant women with FGLS, combined functionally defective genotypes GSTM10 / 0 + GSTT10 / 0 were found in 28.2% of cases (11 pregnant women with FLS) than in the II control group individuals (20.0%), which is 1.4 times higher than in this group. In the group of pregnant women with FGLS “functionally unfavorable” A / G genotypes of the GSTP1 gene was found in 63.04% (29) versus 27.3% [21] of pregnant women without FLS, which was 2.3 times higher than the indicators of this groups (P <0.05). It should be noted that unfavorable homozygous genotypes were detected only in the I - group of pregnant women with FLS, which amounted to 17.4%. As it follows from Table 5, the indicators of the level of specificity and sensitivity of the del / + polymorphism of the GSTT1 gene were SE = 0.8 and SP = 0.75, respectively, at significantly high values (OR = 11.7; 95% CI 5.132- 26.9). At the same time, the calculated AUC indicator demonstrates a high level of effectiveness for predicting the development of the disease, which indicates the possible independent effect of this polymorphism on the risk of pathology development [22-28]. The SE and SP indices of the combined variant of the del polymorphisms of the GSTM1 + GSTT1 genes deviate towards sensitivity and are equal to 0.86 and 0.43, respectively and the efficiency rating is 0.65. These indicators also show a rather significant level of prognostic value of combinations of unfavorable genotypes as a genetic marker for predicting the development of fetal loss syndrome (Figure 2). Then, studies of the expected and observed heterozygous frequencies of the IIe 105 Val polymorphism of the GSTP1 gene in pregnant women with FLS and without revealed distinctive features. A rather significant level of prognostic value of combinations of unfavorable genotypes as a genetic marker for predicting the development of fetal loss syndrome. Then, studies of the expected and observed heterozygous frequencies of the IIe 105 Val polymorphism of the GSTP1 gene in pregnant women with FLS and without revealed distinctive features. (Table 6).

Table 6: The difference between the expected and observed frequencies of heterozygosity of the IIe 105 Val polymorphism of the GSTP1 gene.
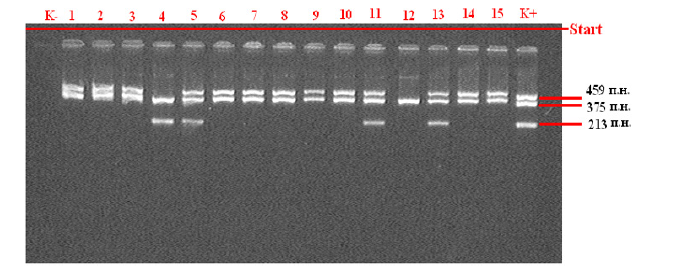
Figure 1: Statistical analysis of the results was carried out using the statistical software package “OpenEpi 2009, Version 2.3”. Note: GSTM1 and GSTT1 gene detection electrophoregram (459 bps - GSTT1 gene, 375 bps - β-globin, 213 bps - GSTM1)
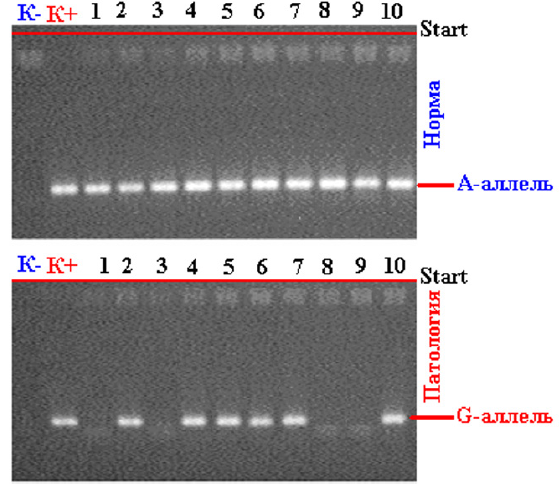
Figure 2: Electrophoregram for the detection of polymorphism (A / G) of the gene mutation -1 glutathione-S-transferase P1 (rs--): K - Negative control; K + Positive control; 1,3,8,9 - wild genotype A/A; 2,4,5,6,7,10 - heterozygous genotype A/G;
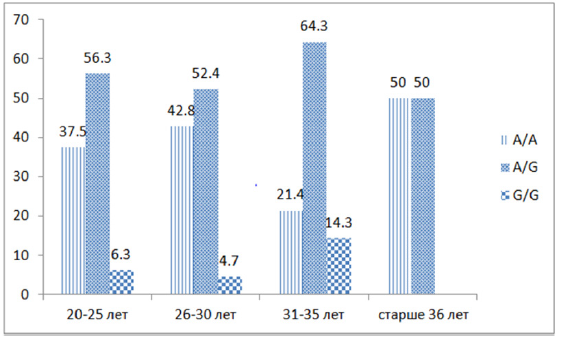
As follows from the figure, the detectability of the association of heterozygous polymorphism of “functionally unfavorable” genotypes A / G IIe 105 Val of the GSTP1 gene was observed in all studied ages - 20–25 years old - in 9 out of 16 (56.3%) pregnant women, 26–30 years old 11 out of 21 (52.4%), 31-35 - 9 out of 14 (64.3%) and over 36 years old 3 out of 6, while homozygous G/G genotypes were found at the age of 20-25 and 26 -30 years- 6.3% and 4.7%, respectively, and at the age of 31-35 years - in 14.3% of cases. Thus, an analysis of the association of intergenic combinations of zero polymorphisms of the GSTM1 and GSTT1 genes (Figure 1) revealed that in the group of pregnant women with fetal loss syndrome, combinations of the homozygous del/del genotype responsible for a lower level of protein product synthesis are significantly more common.
The chance of developing pathology in the presence of this combination of the genotypic variant of del/del genes GSTM1 and GSTT1 significantly increases: up to 7.8 times more compared to other genotypes (χ2 = 12.4; P = 0.0004; OR = 7.8; 95% CI 2.146- 28.65). Whereas, the functionally unfavorable G allele of the GSTP1 gene 2.7 times statistically significantly prevailed in the studied chromosomes of pregnant women with FLS compared with pregnant women without FLS (χ2 = 4.6; P = 0.03; OR = 4.5; 95% CI1.061-19.5). Analysis of the results of molecular genetic studies shows that female individuals of the Uzbek population with combined zero genotypes of the xenobiotic enzymes GSTM1 and GSTT1 (Figure 3), as well as hetero/homozygous genotypes of the IIe 105 Val GSTP1 polymorphism, have a tendency to risk fetal loss syndrome (χ2 = 12.4; P = 0.0004; OR = 7.8; 95% CI 2.146-28.65). Thus, the combined null genotypes GSTM10/0 + GSTT10/0 of the xenobiotic enzyme genes GSTM1 and GSTT1, as well as hetero (G/A) / homozygous (G/G) genotypes of the IIe 105 Val polymorphism of the GSTP1 gene, are significant markers of an increased risk of loss syndrome fetus in Uzbekistan (P <0.05) [29-35]. Allele A and the functionally favorable genotype A/A IIe 105 Val of the GSTP1 gene are significant protective markers for the development of pathology (χ2 = 18.6; P <0.05; OR = 3.9; 95% CI 2.023-7.07). The results obtained indicate that the variants of polymorphisms of the GSTM10/0 + GSTТ10/0 genotypes of the GSTM1 and GSTТ1 genes, as well as the G/A IIe 105 Val genotypes of the GSTP1 gene, are significant prognostic criteria for the risk of fetal growth limit syndrome, which are caused by disorders of the detoxification process in the body in women during pregnancy.
Conclusion
- An analysis of the association of intergenic combinations of zero polymorphisms of the GSTM1 and GSTT1 genes revealed that in the main group of patients, combinations of the homozygous del/del genotype responsible for a lower level of protein product synthesis are significantly more common. The chance of developing pathology in the presence of this combination of the genotypic variant of del/del genes GSTM1 and GSTT1 significantly increases: up to 7.8 times more compared to other genotypes (χ2 = 12.4; P = 0.0004; OR = 7.8; 95% CI 2.146-28.65). there was a slight increase in the frequency of combinations of the heterozygous genotype 0/0 / “+” of these genes in the patient group compared to the control group (60.9% and 52.7%, respectively; χ2 = 0.1; P = 0.3; OR = 1.4; 95% CI 0.697-2.82).
- When analyzing the frequency distribution of alleles and genotypes of this polymorphism in the group of pregnant with FLS, significant differences were found compared with the control group. The functionally unfavorable GST allele of the GSTP1 gene 2.7 times statistically significantly prevailed in the studied chromosomes of pregnant women with FLS compared with pregnant women without FLS (χ2 = 4.6; P = 0.03; OR = 4.5; 95% CI1.061-19.5).
- Thus, the G allele and hetero / homozygous genotypes of the IIe 105 Val polymorphism of the GSTP1 gene are significant markers of an increased risk of developing fetal loss syndrome in Uzbekistan (P <0.05). Allele A and the functionally favorable A / A genotype are significant protective markers for the development of pathology (χ2 = 18.6; P <0.05; OR = 3.9; 95% CI 2.023-7.07).
- Taking into account the age of pregnant women, the detection of “functionally unfavorable” genotypes A / G IIe 105 Val of the GSTP1 gene is observed in women in active reproductive ages - 20–25–36 years 6%, which represents the social significance of the problem.
- Based on the variants of the del/del genotypes of the GSTM1 and GSTT1 genes and G alleles of the GSTP1 gene, one can determine the prognosis of the risk of developing fetal loss syndrome, characterized by a violation of the detoxification process of the body during pregnancy.
- The data obtained allows us to predict the risk of developing fetal loss syndrome, taking into account the assessment of the nature of the detoxification of the body during pregnancy and can be recommended for widespread use of the diagnostic method in obstetric practice.
References
- Altukhov Yu P, Salmenkova EA (2002) Salmenkova E.A. DNA polymorphism in population genetics. Genetics 38(9): 1173-11
- Ahmed-zade VA (2011) Pregnancy and childbirth with antiphospholipid syndrome: course, perinatal outcomes. Medical News 5: 81-85.
- Lyubchich NI, Boboev KT (2015) Study of the role of coagulation system gene polymorphism in the occurrence of preterm birth in women of the Uzbek population. Medical Genetics 14(5): 37-41.
- Gavalov SM, Ryabova OA, Vavilin VA, Lyakhovich VV, Makarova SI (2000) The association of gene polymorphism of xenobiotic biotransformation and detoxification enzymes with the characteristics of bronchial asthma in children. Allergology 3: 14-21.
- Guzov II, Plasmogen activator inhibitor gene polymorphism-1 (PAI-1) risk of developing obstetric pathology
- Gulyamova G Sh, Mavlyanova ShZ, Boboev KT (2015) The role of the polymorphic version of the tumor necrosis factor alpha gene in the development of atopic dermatitis in the population of Uzbekistan. Clinical dermatology and venereology 14(4): 79 -83.
- Melkozyorova OA (2016) Molecular genetic predictors of abnormal uterine bleeding in girls born with intrauterine growth retardation syndrome. Obstetrics and gynecology 95-100.
- NA Davydova, AI Dmitriev, NV Sevostyanova (2007) Analysis of polymorphic variants of the glutathione-S-transferase genes T1, M1 and P1 in patients with prostate cancer. XI Russian Oncological Congress. Congress proceedings 224.
- Baranov VS, Baranova EV, Ivashchenko TE, Aseev MV Petersburg (2000) Human genome and genes of “predisposition”. Introduction to predictive medicine: Intermedika p. 271.
- Bespalova ON (2007) Genetics of miscarriage. Journal of Obstetrics and Women's Diseases 81-95.
- Karimov Hya, Saidov AB, Boboev KT, Assesorova Yu Yu (2001) Fundamental and applied aspects of molecular genetics in medicine. Scientific publication 352.
- Liu SY, Zhang CJ, Si XM, Yao YF, Shi L, et al. (2011) Association between single nucleotide polymorphisms of 5'-untranslated region of GPx4 gene and male infertility 28(3): 270-27
- Popova SN, Slominsky PA, Galushkin SN (2002) Polymorphism of glutathione-8-transferases M1 and T1 in a number of populations of Russia. Genetics 2: 1-4.
- Umarova LN, Ishniyazova ND, Abdurakhmanova FR (2013) Modern ideas about the syndrome of intrauterine growth retardation and the criteria for evaluating it. Bulletin of the Tashkent Medical Academy. 101-105.
- Quick SK, Shields PG, Nie J, Platek ME, Mccann SE, et al. (2008) Effect modification by catalase genotype suggests a role for oxidative stress in the association of hormone replacement therapy with postmenopausal breast cancer risk. Cancer Epidemiol Biomarkers Prev 17(5): 1082-108
- Ramprasath T, Murugan PS, Kalaiarasan E, Gomathi P, Rathinavel A, et al. (2012) Genetic association of Glutathione peroxidase-1 (GPx-1) and NAD(P)H: Quinone Oxidoreductase 1(NQO1) variants and their association of CAD in patients with type-2 diabetes. Mol Cell Biochem 361(1): 143.
- Eliseeva Yu E (2001) Angiotensin-converting enzyme, its physiological role. Questions of medical chemistry 1: 15-21.
- Lyubchich NI (2016) Pathogenetic aspects of preterm labor with thrombophlybia (principles of diagnosis and prevention). Abstract of the doctor74.
- Brain EV (2003) A study of the genetic predisposition to gestosis: polymorphism of genes involved in the regulation of endothelial function. Journal of Obstetrics and Women's Diseases 25-34.
- Spiridonova MG, Trifonova EA, Falyushina SV (2007) Molecular genetic analysis of polymorphic markers of genes responsible for the functioning of factors of the endothelial system in connection with the complicated course of pregnancy. Medical Genetics 7(61): 38-42.
- Aulchenko Yu S, Aksenovich TP (2006) Methodological approaches and strategies for mapping genes that control complex human traits. Bulletin of vogis 10(1): 189-202.
- Baranov VS, Aylamazyan EK (2007) Ecological and genetic causes of reproductive health disorders and their prevention. Journal of Obstetrics and Women's Diseases 1: 3-10.
- Barinova IV (2015) Pathogenesis of antenatal death: phenotypes of fetal loss and thanatogenesis. Russian Bulletin of the Obstetrician-Gynecologist 68-76.
- Makatsaria AD (2007) Antiphospholipid syndrome-immune thrombophilia in obstetrics and gynecology 55–112.
- Melkozyorova OA (2016) Molecular genetic predictors of abnormal uterine bleeding in girls born with intrauterine growth retardation syndrome. Obstetrics and gynecology 95-100.
- Morozova KV, Lutsenko NN (2015) The role of gene polymorphism of antioxidant system enzymes in the genesis of miscarriage. Obstetrics, gynecology and reproduction 54-61.
- Orlov AV, Krukier II, Drukker NA, Kaushanskaya LV (2005) The role of growth factors in the pathogenesis of an undeveloped pregnancy. Russian Bulletin of the Obstetrician-Gynecologist 3: 4-6.
- Tyszkiewicz OB (2004) Features of the hemostatic system in women with habitual miscarriage and the presence in the blood plasma of antiphospholipid antibodies: medical science.
- Fedorova IA, Deryugina LA, Krasnova EI (2012) Molecular genetic aspects of metanephrogenesis in norm and pathology: scientific publication. Russian Bulletin of Perinatology and Pediatrics 57(5): 45-51.
- Khodzhaeva DA, Luninia SN, Lutsenko NN (2011) The role of SOD2 gene polymorphism in women in the genesis of non-developing pregnancy. Bulletin of the Russian State Medical University 2: 134-136
- Seamus J Murphy, Anne E Hughes, Chris C Petterson, Lesley A Anderson, et al. (2007) A population -based association study of SNPs of GSTP1, MnSOD, GPX2 and Barrett, s esophagus and esophageal adenocarcinoma. Carcinogenesis 28(6): 1323-1328.
- Amirchaghmaghi E, Rezaei A, Moini A, Roghaei MA, Hafezi M, et al. (2015) Gene expression analysis of VEGF and its receptors and assessment of its serum level in unexplained recurrent spontaneous abortion. Cell J 4(16): 538-
- Bermano G, Pagmantidis V, Holloway N, Kadri S, Mowat NA, et al. (2007) Evidence that a polymorphism within the 3’UTR of glutathione peroxidase 4 is functional and is associated with susceptibility to colorectal cancer. School of Life Sciences, The Robert Gordon University, Aberdeen, AB25 1HG, UK. Genes Nutr 2(2): 225-32.
- Lin JC, Kuo WR, Chiang FY, Hsiao PJ, Lee KW, et al. (2009) Glutathione peroxidase 3 gene polymorphisms and risk of differentiated thyroid cancer. Surgery 145(5): 508-5
- Mlakar SJ, Osredkar J, Prezelj J, Marc J (2012) Antioxidant enzymes GSR, SOD1, SOD2, and CAT gene variants and bone mineral density values in postmenopausal women: a genetic association analysis. Menopause 19(3): 368-3





 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.