Research Article

 Creative Commons, CC-BY
Creative Commons, CC-BY
Molecular Characterization of Multi Drug Resistance Escherichia coli isolated among Diabetes Mellitus Patients in Dongla State, Sudan
*Corresponding author: Husham M Taha Aloob, Department of Microbiology and Parasitology, Faculty of Medical Laboratory Sciences, Dongla University, Sudan.
Received: June 22, 2020; Published: July 21, 2020
DOI: 10.34297/AJBSR.2020.09.001424
Abstract
Background: Urinary tract infections (UTIs) caused by Escherichia coli have become a significant worldwide public health concern and is a common infectious disease in which level of antimicrobial resistance are alarming worldwide.
Methods: Urine samples were collected form Diabetic patients clinically diagnosed by having UTI, during the period from November 2019 to April 2020 at diabetic center in Dongla, Antibiotic susceptibility tests were performed by Kirby-Bauer disc diffusion and B-lactamases genes were detected used PCR.
Results: A total of 120 E. coli were Isolated from DM with UTI. All the isolated were shown to be resistant to Cefpodoxime (100%). The most efficient antibiotics were Colistin and imipenem (99.2% and 88.3 respectively as susceptibility rate) followed by Gentamycin (70%). High resistance rates were observed with ofloxacin (90.8%), Ciprofloxacin (77.5%), Amikacin (60.8%), ceftriaxone (58.3%) and Cefepime (51.7%). The most B-lactamase genes isolates were blaCTX-M -1 gene (64%) followed by blaCMY-G2 (55.8%), blaSHV gene (34.2%), blaNDM gene (10.8%), blaOXA-48 gene (5.8%), blaVIM gene (3.3%), blaKPC gene (2.5%) and blaIMP were not detected in any of the tested isolates.
Conclusions: Urinary tract infection due to E. coli may be difficult to treat empirically due to high resistance to commonly used antibiotics. Continuous surveillance of multidrug resistant organisms and patterns of drug resistance are needed to prevent treatment failure and reduce selective pressure.
Keywords: UTI; diabetes mellitus; E. coli; Multidrug‑resistant; B-lactamase genes; Sudan
Introduction
Diabetes mellitus is a common endocrine disorder in which there is an insufficiency of, or resistance to, the hormone insulin, which regulates blood glucose levels. It was recorded in 2019 that worldwide 463 million people had diabetes, 90% of which was type II DM. This number is estimated to rise to 552 million in 2030 [1]. The chronic hyperglycemia in diabetes is associated with long-term damage, dysfunction, and failure of various organs, especially the eyes, kidneys, nerves, heart, and blood vessels [2]. DM has long been a predisposing factor for urinary tract infection (UTI) [3]. These factors include weak host immune systems with impaired neutrophil function, depressed T-cell mediated immune response, decreased production of prostaglandin E, thromboxane B2, leukotriene B44 and depressed antioxidant systems [4,5] Worldwide, UTIs’ prevalence was estimated to be around 150 million persons per year [6]. Urinary tract infection can be roughly classified based on anatomy as upper UTI and lower UTI, or both. UTI can be symptomatic or asymptomatic but is defined as presence of bacteriuria with a quantitative count of more than or equal to 105 colony forming unit of bacteria per milliliter [7]. The commonest pathogenic organism isolated in UTI is E. coli followed by K. pneumoniae, Staphylococcus, Proteus, Pseudomonas, Enterococcus, and Enterobacter [8-12]. Escherichia coli is the most common causative agent of UTIs in both DM and non-DM patients [11]. Uropathogenic E. coli (UPEC) possesses a variety of pathogenicity determinants that make colonization of the urinary tract possible. These determinants include fimbrial (P, S/F1C and type 1 fimbriae) and non-fimbrial adhesins that mediate bacterial adherence to epithelial cells, siderophores (iron-acquisition systems), secreted toxins (haemolysin and cytotoxic necrotizing factor 1) and capsule forming polysaccharides for immune evasion [13-15]. Phylogenetic analysis classifies E. coli strains into four main groups (A, B1, B2 and D). Groups B2 and D are mainly associated with E. coli strains causing extraintestinal infections, whilst groups A and B1 are associated with commensal strains [16]. Worldwide, there is great concern due to the high rates of resistance to antimicrobials used in the treatment of infections caused by E. coli, particularly in developing countries. Frequent prescription of antibiotics, including the ones with broad-spectrum, may result in development of antibiotic-resistant urinary pathogens. Since patients with DM are more prone to have resistant pathogens, they inevitably require longer and more potent antimicrobial treatment [17]. Therefore, improved control of glycaemia in diabetics may help in controlling the UTIs. Accurate screening for UTI in diabetic patients is also critical to enable the appropriate treatment, avoiding related complications. In this study we aimed to assess the prevalence of E. coli and pattern of the antimicrobial drugs susceptibility by phenotyping and genotyping method.
Material and Methods
Study design
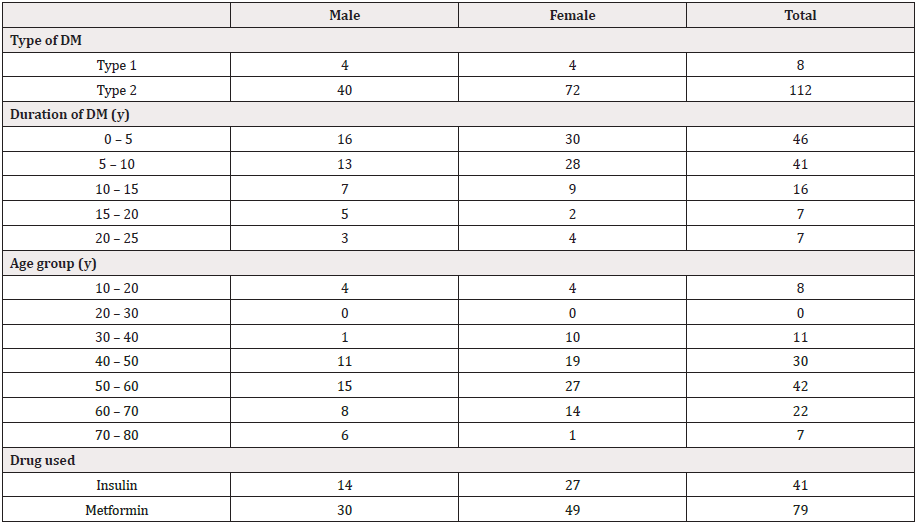
A cross-sectional study was carrying out during the period from August 2019 to April 2020 at diabetic center in Dongla and Neelain university. The study includes patients clinically diagnosed by having one or more of the following symptoms: dysuria, frequency, urgency, suprapubic discomfort, or flank pain. Non-diabetic and pregnancy were excluded from the study. A total of 120 E. coli isolated form Diabetic patients (45 males and 75 females) with age group ranged from 10 to 80 years old. All patients were informed of the purpose of the study and their consent or that of their care provider was obtained before urine samples were collected.
Sample collection and Processing
Each patient was asked to collect approximately 10-20 ml of midstream urine into a sterile urine container. after giving proper instructions to avoid contamination and samples were processed in the laboratory within 2 hours of collection. None of the patients admitted to consuming antibiotics during the 2 weeks prior to urine sample collection.
Data collection
A structured questionnaire and referring to the patient clinical sheet were being used; demographic data and other Data (clinical symptoms, previous antibiotic, duration of antibiotic used). verbal consent was obtained from each patient enrolled in this study.
Isolation and identification of Escherichia Coli using Biochemical tests and selective medium
Urine cultures were performed using semi-quantitative technique whereby urine samples were inoculated on cystein-Lactose electrolyte deficient (CLED) medium plates with a calibrated loop (0.001ml) and incubated at 37°C for 18-24 hours. Urine culture reports that exhibited colony forming units (CFUs) more than 105/ ml of voided urine were considered significant [18]. Isolated colonies from significant plates were identified and differentiated from related organisms using standard conventional biochemical tests (Kligler Iron agar: slant /Acid, butt/ Acid, H2S production / -, Gas / +; Motility test / motile; Indole /+, Urease / - ; Citrate /-) according to [19].
Antimicrobial susceptibility testing
Antimicrobial sensitivity testing of all isolates was performed on diagnostic sensitivity test plates according to the Kirby-Bauer method [20] following the definition of the Committee of Clinical Laboratory International Standards [21]. Bacterial inoculums were prepared by suspending the freshly grown bacteria in 5mL sterile saline. A sterile cotton swab was used to streak the surface of Mueller Hinton agar plates. Filter paper disks containing a designated concentration of the antimicrobial drugs were obtained from Hi-Media Laboratories in the following concentrations: Amikacin (30μg), Gentamycin (10μg), Cefotaxime (30μg), ceftriaxone (30μg), Meropenem (10μg) ciprofloxacin (5μg), ofloxacin (5μg), colistin (10μg), Cefepime (30μg). The diameters of zone of inhibition were interpreted according to CLSI standards. Media and disks were tested for quality control with E. coli standard strain.
Molecular detection
DNA for molecular detection was extracted after bacterial lysis according to the extraction protocol prepared by the Community Reference Laboratory for Antimicrobial Resistance (CRL, 2009). Briefly, a few colonies taken from fresh culture medium and transferred to phosphate buffered saline (pH 7.3). The suspension was then heated at 100°C for 15 minutes. Boiled suspension was transferred directly on ice, this was followed by vortexing and the suspension was then centrifuged at 12000 rpm for 5 minutes to sediment the debris, the clear supernatant was used as template DNA in PCR method.
The universal primers were designed for b-lactamase genes, including ESBL genes (blaSHV and blaCTX-M-1), AmpC genes (blaCMY- G2), carbapenemases genes (blaKPC, blaIMP, blaVIM, blaNDM and bla- OXA-48). Details for the primers were shown in (Table 1). PCR was performed and the test was carried out in total volume of 25 μl, containing 5 μl master mix (Solis Bio dyne master mix), 2 μl of primer, 5 μl of DNA and 13 μl of distilled water. The PCR was performed for 35 cycles (initial denaturation at 95 oC for 5 min, denaturation at 95 oC for 50 s, annealing at 56 oC for 40 s or 60°C for 45, elongation at 72 oC for 1 min for 35 cycles and final extension at 72 oC for 5 min). 5 μl of the PCR product was analyzed using 1% or 1.5% Agarose gel electrophoresis and stained with 0.15% Ethidium bromide and the product was visualized using UV gel documentation [22,23].
Data analysis
Statistical analysis was done by using Statistical Package for Social Science program (version 20).
Results
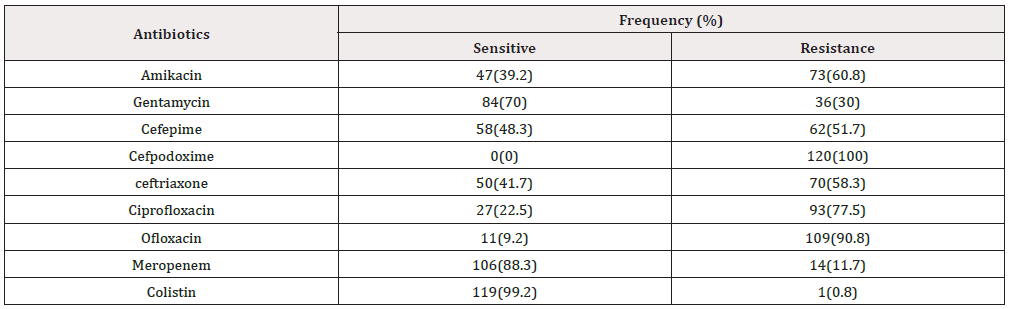
total of 120 isolates were identified as E. coli from DM with UTI (45 males and 75 females, 6.7% had type 1 and 93.3% had type II DM with age group ranged from 10 to 80 years old), by routine phenotypic methods including Gram’s staining, colony morphology and standard conventional biochemical tests. About 25 % and 35% of the UTI positive patients were in age between 40-50 and 50-60 years, respectively (Table 2). This result indicated that the emergence of UTI raised with the increase in age of the patient. E. coli strains showed differences in susceptibility and resistance patterns to the antimicrobials tested. All of the isolated uropathogenic E. coli were shown to be resistant to Cefpodoxime (100%). The most efficient antibiotics were Colistin and imipenem (99.2% and 88.3 respectively as susceptibility rate) followed by Gentamycin (70%). High resistance rates were observed with ofloxacin (90.8%), Ciprofloxacin (77.5%), Amikacin (60.8%) and ceftriaxone (58.3%). Resistance to Cefepime (51.7%) was a high rate of intermediate resistance (Table 3). Regarding the MDR isolates, all of E. coli isolates collected for this study showed resistance to four or more antibiotics.
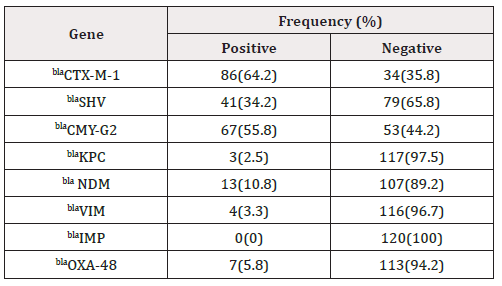
By applying PCR method among the 120 E. coli isolates were tested for the production of B-lactamase genes, including ESBL genes (blaSHV and blaCTX-M-1), AmpC genes (blaCMY-G2), carbapenemases genes (blaKPC, blaIMP, blaVIM, blaNDM and blaOXA-48) (Figure 1-4). The most B-lactamase genes isolates were blaCTX-M -1 gene (64%) followed by blaCMY-G2 (55.8%), blaSHV gene (34.2%), blaNDM gene (10.8%), blaOXA-48 gene (5.8%), blaVIM gene (3.3%), blaKPC gene (2.5%) and blaIMP were not detected in any of the tested isolates (Table 4).
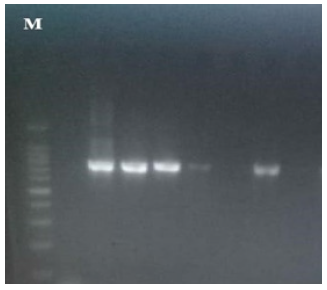
Figure 1: blaCMY-G2, blaCTX-M-1, blaKPC genes DNA results (963bp, 944bp and 900bp respectively) on 1% agarose gel. Lane M shows 100 bp DNA marker, lane 1 shows negative control, lanes 3 show positive results for blaCMY-G2, lanes 6 and 7 show positive results for blaCTX-M-1, lanes 8 and 9 show positive results for blaKPC, lane 2,4,5,10,11,12 shows negative results.
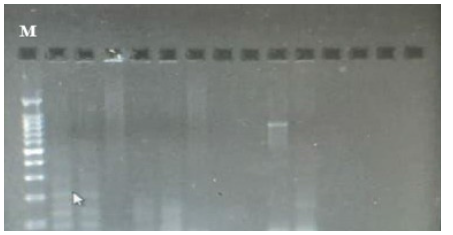
Figure 2: blaSHV gene DNA results (896bp) on 1% agarose gel. Lane M shows 100 bp DNA marker, lane 1 shows negative control, lane 9 shows positive results, lanes 2,3,4,5,6,7,8,10,11,12,13 and 14 show negative results.
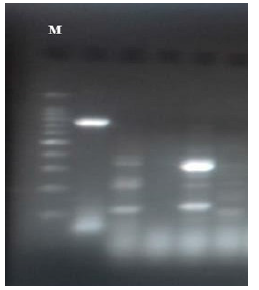
Figure 3: blaOXA-48 and blaVIM DNA results (743bp and 389bp respectively) on 1.5% agarose gel. Lane M shows 100 bp DNA marker, lane 1 shows show positive results for blaOXA-48. lane 2 and 4 shows positive results for blaVIM. lanes 2 and 5 show negative results.
Discussion
Urinary tract infections (UTIs) are the most common human infectious disease affecting the bladder, kidneys and urinary tracts [24]. Kidney stones, diabetes, weak immune system can increase the risk of UTIs [25]. Patients with significant bacteriuria have at least two symptoms referable to the urinary tract infection (dysuria, urgency, frequency, incontinence, suprapubic pain, flank pain or costovertebral angle tenderness, fever (temp ≥ 38°C) and chills are said to be symptomatic. Complications of UTI include urosepsis, renal impairment, and renal abscess [26,27]. In the present study all urine sample (100%) revealed significant growth. the most isolated agents from urinary tract infections vary, almost all of them are caused by single microorganism type. In this study the most frequently isolated microorganism was E. coli with a rate of (75%). this result is agreement with findings of previous studies [28-33]. Uropathogenic Escherichia coli (UPEC), is one of the main causes of community (80–90%) and nosocomial acquired UTIs (30–50%) [34-37]. Also, our results reveal higher prevalence of urinary tract infections in female patients than in male [38-40]. this mainly due to short urethra, absence of prostatic secretion, pregnancy, and easy contamination of the urinary tract with fecal flora [41]. The emergence of high rates of antibiotic resistance and MDR-phenotype from urinary tract infections related bacteria becomes a public health concern worldwide. Our results findings that E. coli isolated were showed the following resistance rates; Cefpodoxime (100%), floxacin (90.8%), Ciprofloxacin (77.5%), Amikacin (60.8%) ceftriaxone (58.3%), Cefepime (51.7%), imipenem (11.7%), Colistin (0.8%). These results agreed with results of [42-46]. There is an increase in resistance of Uropathogens to most antibiotics because of excessive and inappropriate usage, reducing. These findings are clearly alarming as our country could be running out of effective antibiotics if this trend continues.
Current studies mainly focus on a variety of function enzymes produced in E. coli, including ESBLs, plasmid mediated AmpCs, and carbapenemases. Global emergence and spread of carbapenemase genes and ESBL genes among E. coli isolates, poses severe challenges to public health. Furthermore, this study indicated that ESBL positivity was closely related to the resistance of most drugs. In recent years, multidrug resistant caused by ESBLs are reported to be associated with higher morbidity and mortality rates [48]. The proportion of the ESBL positive cases was highest, followed by AmpC-producing stains, and carbapenemases-producing stains. ESBLs are mainly mediated by plasmid, while AmpCs are mainly mediated by chromosome. CTX-M types are the major phenotypes of domestic ESBLs, Widespread dissemination of these genes has been described in Africa and elsewhere, followed by SHV type [49]. AmpC genotype is given priority to blaCMY in the worldwide, especially the subtype of blaCMY-G2 [50]. In the present study, prevalence of 𝛽-lactamases producing isolates were found in (Table 4). Resistance to carbapenems can be acquired through mechanisms such as drug efflux, loss of porins, and carbapenemase-production [51], the latter of which is predominantly caused by the serine-carbapenemases such as K. pneumoniae carbapenemase (KPC) and oxacillinase -lactamase (OXA), or metallo- lactamases including Verona integron-encoded metallo- -lactamase (VIM), New Delhi metallo- -lactamase(NDM), or imipenem as emetallo- -lactamase(IMP) [52]. New Delhi metallo-β-lactamase (NDM) and carbapenem-hydrolyzing oxacillinase-48 (OXA-48) are the most common carbapenemases among E. coli worldwide [53]. Metallo-lactamase-producing E. coli isolates were showed prevalence rates of 10.8%, 5.8%, 3.3 % and 2.5% for bla NDM, blaOXA-48, blaVIM and blaKPC, respectively; In this study, no blaIMP genes was found, but we cannot exclude that other MBL genes might have been involved in resistance.
Furthermore, reports of emerging (VIM, NDM, KPC and IPM,) MBL genes among Gram-negative strains have been published worldwide, including sub-Saharan Africa, Egypt [54], Uganda [55], Ethiopia [56], Kenya [57], South Africa [58], Saudi Arabia [59]. Thus, other carbapenemase-encoding genes should be evaluated in future studies and PCR should be conducted for detecting all carbapenemase-encoding genes in carbapenem resistant isolates. selecting antibiotics for treating bacterial infections should be according to the culture and sensitivity results and the international guidelines to minimize further MDR development, cost-related and health-related consequences.
Conclusions
In this study, a high prevalence of resistance to β-lactams as well as to other antimicrobials were observed in E. coli isolates from DM patients with UTI. The study showed the importance of continuous monitoring programming of multidrug resistance in our hospitals. It also showed the need for developing attempts to decrease the prevalence of ESBL producing organisms and the modification of guidelines for UTIs. ESBLs are clinically significant and patients infected with ESBL-producing Enterobacteriaceae experience a greater likelihood of poor outcome if they are treated with inappropriate antibiotics. The exception is uncomplicated urinary tract infections where a very high urinary concentration of β-lactam antibiotics can be achieved.
References
- International Diabetes Federation (2011) IDF Diabetes Atlas, Belgium.
- Bonadio M, CostarelliS, Morelli G, Tartaglia T (2006) the influence of diabetes mellitus on the spectrum of uropathogens and the antimicrobial resistance in elderly adult patients with urinary tract infection. BMC infectious diseases 6(1): 54.
- Sahib AKY (2008) Study of ciprofloxacin resistant Escherichia coli (CREC) in type 2 diabetic patients with symptomatic urinary tract infections. Iraq J Comm Med 21(1): 58-63.
- Boyko EJ, Fihn SD, Scholes D (2005) Risk of urinary tract infection and asymptomatic bacteriuria among diabetic and nondiabetic postmenopausal women. Am J Epidemiol 161(6): 557-564.
- Joshi N, Caputo GM, Weitekamp MR (1999) Infections in patients with diabetes mellitus. N Engl J Med 341(25): 1906-1912.
- K Gupta, DF Sahm, D Mayfield, WE Stamm (2001) “Antimicrobial resistance among uropathogens that because community acquired urinary tract infections in women: a nationwide analysis,” Clinical Infectious Diseases 33(1): 89-94.
- Flores Mireles, Ana L (2015) Urinary Tract Infections: Epidemiology, Mechanisms of Infection and Treatment Options. Nature reviews Microbiology 13(5): 269-284.
- AR Manges, P Natarajan, OD Solberg, PS Dietrich, LW Riley, et al. (2006) “The changing prevalence of drugresistant Enterobacteriaceae groups in a community: evidence for community outbreaks of urinary tract infections.” Epidemiology and Infections 134(2): 425-431.
- EAE, KIO (2008) “Incidence and antibiotic susceptibility pattern of Staphylococcus aureus amongst patients with urinary tract infection (UTI) in UBTH Benin City, Nigeria.” African Journal of Biotechnology 7(11): 1637-1640.
- Nielubowicz GR, Mobley HL (2010) Host-pathogen interactions in urinary tract infection. Nat Rev Urol 7(8): 430-441.
- Bonadio M, Costarelli S, Morelli G, Tartaglia T (2006) The influence of diabetes mellitus on thespectrum of uropathogens and the antimicrobial resistance in elderly adult patients with urinary tract infection. BMC Infect 6: 54.
- Bonadio M, Meini M, Spitaleri P, Gigli C (2001) Current microbiological and clinical aspects of urinary tract infections. Eur Urol 40(4): 439-445.
- Bower JM, Eto DS, Mulvey MA (2005) Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic 6(1): 18-31.
- Tseng CC, Huang JJ, KoWC, Yan JJ, Wu JJ, et al. (2001) Decreased predominance of pap G class II allele in Escherichia coli strains isolated from adults with acute pyelonephritis and urinary tract abnormality. J Urol 166(5): 1643-1646.
- Wiselka MF (1996) Bacterial and host factors in the pathogenesis of urinary tract infections. In Infections of the Kidney and Urinary Tract, Edited by WR Cattell. New York: Oxford pp. 8-33.
- Picard B, Garcia JS, Gouriou z, Duriez P, Brahimi N, et al. (1999) The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67(2): 546-553.
- Schneeberger C, Stolk R, DeVries H (2008) Differences in the Pattern of Antibiotic Prescription Profile and Recurrence Rate for Possible Urinary Tract Infections in Women with and Without Diabetes. Diabetes Care 31(7): 1380-1385.
- Wilson ML, Gaido L (2004) Laboratory diagnosis of urinary tract infections in adult patients. Clin Infect Dis 38(8): 1150-1158.
- McCartney JE, Collee JG, Mackie TJ (1989) Practical medical microbiology. Charchil Livingstone.
- AW Bauer, WM Kirby, JC Sherris, M Truck (1966) “Antibiotic susceptibility testing by a standardized single disk method,” American Journal of Clinical Pathology 45(4): 493-496.
- Clinical and Laboratory Standards Institute [CLSI]. “Performance Standards for Antimicrobial Susceptibility Testing- Approved Standard”. 11th edition. M02-A11. 32.1 (2012).
- Diana D, Gisele P, Christine L, Tracie L, Deirdre L, et al.(2012)Laboratory detection of Enterobacteriaceae that produce carbapenemases." Journal of clinical microbiology 50 (12): 3877-3880.
- Du J, Li P, Liu H, Lü D, Liang H, et al. (2014) Phenotypic and molecular characterization of multidrug resistant Klebsiella pneumoniae isolated from a university teaching hospital, China. PloS one 9(4).
- Tarchouna M, Ferjani A, Ben Selma W, Boukadida J (2013) Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. International Journal of Infectious Diseases 17(6): e450-e453.
- Emody L, Kerenyi M, Nagy G (2003) Virulence factors of uropathogenic Escherichia coli. Int J Antimicrob Agents 22: 29-33.
- Loh K, Sivalingam N (2007) Urinary tract infections in pregnancy. Malays FAM Phys 2: 54-59.
- Badri AM, Mohamed SG (2017) Clinical Epidemiology and Antibiogram of UTI Patients Attended Different Hospital in Khartoum. Sudan Clin Microbial 6(5): 301.
- Amin M, Mehdinejad M, Pourdangchi Z (2011) Study of bacteria isolated from urinary tract infections and determination of their susceptibility to antibiotics. Jundishapur J Microbiol 2: 118-123.
- Beyene G, Tsegaye W (2011) Bacterial Uropathogens in Urinary Tract Infection and Antibiotic Susceptibility Pattern in Jimma University Specialized Hospital, Southwest Ethiopia. Ethiop J Health Sci 21(2): 141-146.
- Machado Alba JE, Murillo Muñoz MM (2012) Evaluación de sensibilidad antibiótica en urocultivos de pacientes en primer nivel de atención en salud de Pereira. Rev Salud Publica (Bogota) 14(4): 710-719.
- Al Yousef SA (2016) Clinical and laboratory profile of urinary tract infections associated with extended spectrum beta-lactamase producing Escherichia coli and Klebsiella pneumoniae. Ann Clin Lab Sci 46(4): 393-400.
- Seija V, Frantchez V, Pintos M (2010) Etiología de la infección urinaria de adquisición comunitaria y perfil de susceptible de Escherichia coli a los principals’ agents antimicrobianos. Rev Med Urug (Montev) 26: 14-24.
- Al Otaibi FE, Bukhari EE (2013) Clinical and laboratory profiles of urinary tract infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in a tertiary care center in central Saudi Arabia. Saudi Med J 34(2): 171-176.
- Kucheria R, Dasgupta P, Sacks SH, Khan MS, Sheerin NS, et al. (2005) Urinary tract infections: new insights into a common problem. Postgrad Med J 81(952): 83-86.
- Hooton TM (2012) Uncomplicated urinary tract N Engl J Med 366: 1028-1037.
- Laupland KB, Ross T, Pitout JD, Church DL, Gregson DB, et al. (2007) Community-onset urinary tract infections: a population-based Infection 35(3): 150-153.
- Nicolle LE(2001) A practical guide to the management of complicated urinary tract Drugs Aging 18(4): 243-254.
- RD Harrington, TM Hooton (2003) “Urinary tract infection risk factors and gender,” The Journal of Gender-Specific Medicine 3(8): 27-34.
- Bouchillon S, Hoban DJ, Badal R, Hawser S (2012) Fluoroquinolone resistance among gram‑negative urinary tract pathogens: global smart program results, 2009–2010. Open Microbiol J 6: 74-78.
- Ali I, Rafaque S, Ahmed S, Malik JI, Dasti P, et al. (2016) Prevalence of multi‑drug resist‑ ant uropathogenic Escherichia coli in Potohar region of Pakistan. Asian Pac J Trop Biomed 6(1): 60-66.
- Haider G, Zehra N, Afroze A, Haider A (2010) Risk factors of urinary tract infection in pregnancy. J Pak Med Assoc 60(3): 213-216.
- Parajuli NP, Acharya SP, Mishra SK, Parajuli K, Rijal BP, et al. (2017) High burden of antimicrobial resistance among gram negative bacteria causing healthcare associated infections in a critical care unit of Nepal. Antimicrob Resis Infect Contr 6: 2-9.
- Ouno GA, Korir SC, Cheruiyot JC, Ratemo OD, Mabeya BM, et al. (2013) Isolation, Identification and Characterization of Urinary Tract Infectious Bacteria and the Effect of Different Antibiotics. J Nat Sci Res 3: 150-159.
- Leoni AF, Monterisi A, Acuña PG (2017) Community acquired urinary tract infections in older adults. Rev Fac Cien Med Univ Nac Cordoba 74(1): 10-17.
- Fasugba O, Mitchell BG, Mnatzaganian G, Das A, Collignon P, et al. (2016) five-year antimicrobial resistance patterns of urinary Escherichia coli at an Australian tertiary hospital: time series analyses of prevalence data. PLoS One 11(10): e0164306.
- Ramírez Castillo FY, Moreno Flores AC, Avelar González FJ , Márquez Díaz F , Harel J, et al.(2018) An evaluation of multidrug-resistant Escherichia coli isolates in urinary tract infections from Aguascalientes, Mexico: cross-sectional study. Annals of clinical microbiology and antimicrobials. 17(1): 34.
- Marchaim D, Gottesman T, Schwartz O, Korem M, Maor Y, et al. (2010) National multicenter study of predictors and outcomes of bacteremia upon hospital admission caused by Enterobacteriaceae producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother 54(12): 5099-5104.
- Younes A, Hamouda A, Dave J, Amyes SG (2011) Prevalence of transferable blaCTX-M-15 from hospital- and community-acquired Klebsiella pneumoniae isolates in Scotland. J Antimicrob Chemother 66(2): 313-318.
- Turner MS, Andersson P, Bell JM, Turnidge JD, Harris T, et al. (2009) Plasmidborne blaSHV genes in Klebsiella pneumoniae are associated with strong promoters. J Antimicrob Chemother 64(5): 960-964.
- Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, et al. (2008) Dissemination of clonally related Escherichia coli strains expressing extended-spectrum betalactamase CTX-M-15. Emerg Infect Dis 14 (2): 195-200.
- Fernández L, Hancock RE (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25(4): 661-681.
- Tascini C, Tagliaferri E, Giani T, Leonildi A, Flammini S, et al.(2013) Synergistic activity of colistin plus rifampin against colistin-resistant KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother AAC 57(8): 3990-3993.
- Nordmann P, Poirel L (2014) the difficult-to-control spread of carbapenemase producers in Enterobacteriaceae worldwide. Clin Microbiol Infect 20(9): 821-830.
- Diab M, Fan N, El Said M, El Dabaa E, El Defrawy I, et al. (2013) Occurrence of VIM-2 Metallo-β-lactamases in imipenem resistant and susceptible Pseudomonas aeruginosa clinical isolates from Egypt. Afri J Microbial 7(35): 4465-4472.
- Zafer MM, Al Agamy NH, El Mahallawy HA, Amin MA, Ashour M, et al. (2014) Antimicrobial resistance pattern and their β-lactamase encoding genes among Pseudomonas aeruginosa strains isolated from cancer patients. Bio Med Research Int e101635.
- Pritsch M, Zeynudin A, Messerer M, Baumer S, Liegl G, et al. (2017) First report on blaNDM-1-producing Acinetobacter baummanii in three clinical isolates from Ethiopia. BMC Infect Dis 17: 180.
- Poirel L, Revathi G, Bernabeu S, Nordmann P (2011) Detection of NDM-1producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother 55(2): 934-936.
- Sigh Moodley A, Perovic O (2016) Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enterobacteriaceae in South Africa. BMC Infect Dis 16(1): 536.
- Shaik S, Fatima J, Shakill S, Rizvi SMD (2015) Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology, and treatment. Saudi Biol Sci 22(1): 90-101.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.