Opinion 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Novel Strategies to Tackle Ras-Related Cancer: An Opinion Based on Two Recent Reviews
*Corresponding author: Paul D Adams, Department of Chemistry and Biochemistry, Program in Cellular and Molecular Biology, University of Arkansas, Fayetteville, USA.
Received: April 30, 2020; Published: June 18, 2020
DOI: 10.34297/AJBSR.2020.09.001376
Opinion
In this communication, we provide a brief overview of two recent reviews by [1,2] which highlight the importance of the continued need for the development of small molecule inhibitors to modulate abnormal signaling processes stimulated by the Ras GTPases. If not properly regulated, these signaling processes can result in Ras-stimulated hyperactivity that can lead to diseases such as cancer. However, to date, strategies to target inhibition of these proteins still remains a daunting challenge. Therefore, these proteins remain important targets for drug discovery efforts to control Ras-stimulated abnormal cell signaling.
Evidence continues to show that members of the Ras-related GTPases undergo deregulated mechanisms that can lead to cancer. These proteins are involved in several cell signaling activities that are vital to processes such as cell proliferation and differentiation that, if not properly regulated, can result in an oncogenic state [1]. This issue underlies their importance as targets for drug discovery efforts to control Ras-stimulated abnormal cell signaling. Recent reviews by [2], as well as Prieto Dominguez, Parnell & Teng [1] have highlighted the importance of small molecule drug targeting to modulate abnormal Ras-stimulated signaling processes. This protein family serves as binary molecular switches that cycle between active- (GTP-linked) and inactive- (GDP-linked) bound states that help to regulate numerous cellular processes. These small GTPases exhibit intrinsic albeit slow GTP hydrolysis activity and therefore exist predominantly in vivo in the GDP linked inactive state until activated by guanine-nucleotide exchange factors (GEFs), which replaces the bound GDP with GTP. In addition, the enzymes interact with GTPase-activating proteins (GAPs) which cause deactivation by stimulating the hydrolysis of GTP to GDP ([1] and references therein).
The noted reviews discuss recent approaches/mechanisms towards the inhibition of Ras-related proteins in various cancers caused by overexpression, mutation(s), hyperactivity, or undesired Protein-Protein Interactions (PPIs). Designing and identifying particular small molecule inhibitors to decrease and/or eliminate abnormal cell signaling would represent a major step forward in cancer therapies (Figure 1). Target inhibitors of the Ras-related protein ADP ribosylation factor 1 (Arf1) include LM11, Exo2, Brefeldin A (BFA), AMF-26 which function to alter the making of the Arf1-GEF assembly by hampering the GEF activity as described by [1] and references therein). The small molecule Sec7 inhibitor H3 (SecinH3), a non-specific inhibitor of Arf, was described as being able to block signaling activities of Arf1 and Arf6 through binding to the ARF Nucleotide-Binding site Opener (ARNO) Sec7 catalytic domain and thereby deactivating small ARF-specific GEFs [3]. Moreover, this review also highlighted the use of an RNA aptamer (M69), and its ability to hamper Arf actions by deactivating GEFs by binding to the catalytic Sec7 domain [4].
Recent targets towards the inhibition of Ras in cancer have continued to focus on the Ras members H-RAS, N-RAS and K-RAS, which are among the most mutated in human cancers. This emphasis has prompted the diligence and urgency needed in regard to the design and development of small molecule inhibitors that can target these Ras proteins. Despite diligent efforts, there is so far no efficient Ras inhibitor being used to treat cancer [5]. In both reviews, the authors highlight recent experimental approaches to inhibit Ras oncogenic activities. Among the promising approaches is the targeting of small molecules to influence effector interactions. It is very plausible to consider inhibiting Ras-related small GTPase interactions with effectors such as GEFs, which could help to stabilize the inactive state of a Ras-related protein to reduce potential hyperactivity. The two reviews also highlighted some new possibilities to inhibit Ras oncogenic activities, through the use of library screenings, including Bisphenol A and its derivatives 4,4’-biphenol, SCH-53870 and SAH-SOS1 [6] as well as SCH-54292 [7] which targeted NIH3T3 cell proliferation in an apoptoticdependent manner. [2] also cited metal-cyclens, organometallic molecules containing bivalent metals such as Zn2+, Co2+ or Cu2+, which were shown to exert potential inhibitory impact on Ras oncogenic activities [8].
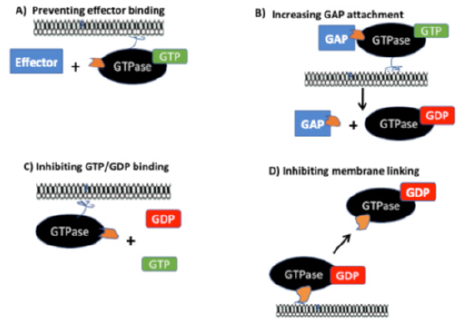
Figure 1: Approaches to offset small GTPase interactions in human cancers: A.GTPase-Effector interactions interrupted via attachment of a small molecule inhibitor (SMI) to the GTPase B.GTPase interactions with GAPs increased by a SMI to enhance GTP conversion to GDP C.GTPase interactions with the SMI to inhibit the binding of either GTP or GDP D.Prevention of membrane attachment for the small GTPase by SMI binding to the protein.
The reviews also pointed out another strategy to tackle Ras oncogenic activities by offsetting interactions that are crucial in Ras signaling transmission mechanisms. Compounds such as enantiomeric iridium (III), R11.1.6, NS1, Sulindac sulfide, indomethacin, MCP1, MCP53, and MCP110 were showcased as having sufficient efficacy to hinder Ras-Raf (Rapidly accelerated fibrosarcoma proteins that are upstream effectors of Ras) interactions thereby frustrating cancer cell proliferation and decreasing tumor size in many cancers ([2] and references therein). Moreover, the reviews described approaches to combat Ras oncogenic activities by stimulating activities of GAPs specific to Ras. Several Ras-specific GAPs inducers were described, such as, RGMA (repulsive guidance molecule A) and Sema4D (Semaphorin4D) which have shown promising results in the stimulation of Rasspecific GAPs to aid in the inactivation of Ras over-activity ([2] and references therein). Moreover, targeting Ras-related proteins to inhibit their attachment to the cell membrane, thereby reducing their activity has also been shown to be a promising approach. The small molecules Salirasib (transfarnesylthiosalicylic acid) and Secramine A were highlighted as having inhibitory effects on small GTPase attachment thereby influencing their hyperactive potential ([1,2] and references therein). It is also worth mentioning that, in addition to highlighting demonstrations of promising results both in vivo an in vitro in the design and development of more potent chemical Ras proteins inhibitors, the reviews also pointed out the potential of non-chemical strategies.
The design of killer genes with the potential of knocking down Ras genes essential for tumerigenesis, as noted by [5], was suggested as an alternative approach to suppress Ras protein activity given issues related to developing potent and effective “Direct” Ras inhibitors. The inhibition of sub-family members of Ras is also an important approach to combat hyperactivity leading to the oncogenic state. Both reviews by [1,2], and references therein, pointed out that developing inhibitory molecules that might target Ras homology family member A proteins, known as RhoA, are important in order to try and minimize tumor development via the stimulation of cell growth, angiogenesis, migration and metastasis in cancerous cells caused by RhoA-stimulated hyperactivity. To that end, RhoA inhibitors like Rhosin, Y16, and CHS-111, have shown promising results in treating Rho GTPase-stimulated hyperactivity [9]. Also highlighted was the ability of Rab proteins to become involved in cancer development by promoting cell growth, invasive behavior through the stimulation of cell cycle continuation, migration and growth, in particular through the use of Rab geranylgeranyl transferase (RabGGTase) inhibitors [10].
Of particular importance to the research efforts of the authors of this opinion, the reviews of [1,2] highlight the importance of design and development of inhibitors of the Ras-Related protein Cdc42 as a great approach in the treatment of many cancers as Cdc42 is known to be involved in the:
a) Promotion of growth of transformed cancer cells by stimulating cell cycle continuance,
b) Induction of tumor formation via the impediment of EGFR breakdown by ubiquitin-proteasome system, and
c) Promotion of migration, invasion and EMT.
Cdc42 inhibitors such as Secramine A, which offsets the attachment of the prenylated Cdc42 to the cell membrane through inhibition of Rho guanine dissociation inhibitor 1, ZCL278, which has been shown to prevent binding between Cdc42 and Intersection (ITSN) in vivo, a GEF which stimulates Cdc42 activity, AZA1 (a structural derivative of NSC23766), and R-ketorolac, an enantiomer of the analgesic S-ketorolac, were all highlighted in terms of their potential in inhibiting Cdc42-stimulated abnormal cell-signaling processes. It is of note that the influence of ZCL278 and AZA1 derivatives on Cdcd42 interactions is currently being studied in the laboratory of the authors of this opinion.
Currently, there are no small GTPase inhibitors used in clinical treatments to address the abnormal activity of these proteins. Although these proteins contribute to several types of cancers, issues related to finding surface areas to serve as binding sites for these small molecule inhibitors still remain a hurdle. Indeed, there is still much work needed in the development of suitable inhibitors to treat small GTPase-stimulated cancers with great efficacy. Nevertheless, the reviews of [1,2] have showcased several promising developments in this regard.
Acknowledgements
This publication was supported by grants from the Arkansas Biosciences Institute, the Arkansas Breast Cancer Research Program and the University of Arkansas for Medical Sciences.
References
- Prieto-Dominguez N, Parnell C, Teng Y (2019) Drugging the Small GTPase Pathways in Cancer Treatment: Promises and Challenges. Cells 8(3): 255.
- Maldonado MDM, Dharmawardhane S (2018) Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res 78(12): 3101-3111.
- Hafner M, Schmitz A, Grune I, Srivatsan SG, Paul B, et al. (2006) Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature 444(7121): 941-944.
- Germer K, Leonard M, Zhang X (2013) RNA aptamers and their therapeutic and diagnostic applications. Int J Biochem Mol Biol 4(1): 27-40.
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ, et al. (2014) Drugging the undruggable RAS: Mission possible? N Nature Reviews Drug Discovery 13(11): 828-851.
- Khan I, Spencer-Smith R, O Bryan JP (2019) Targeting the alpha4-alpha5 dimerization interface of K-RAS inhibits tumor formation in vivo. Oncogene 38(16): 2984-2993.
- Wang W, Fang G, Rudolph J (2012) Ras inhibition via direct Ras binding--is there a path forward?. Bioorganic & Medicinal Chemistry Letters 22(18): 5766-5776.
- Rosnizeck IC, Graf T, Spoerner M, Trankle J, Filchtinski D, et al. (2010) Herrmann C, Gremer L, Vetter IR, Wittinghofer A, Konig B, Kalbitzer HR. Stabilizing a weak binding state for effectors in the human ras protein by cyclen complexes. Angew Chem Int Ed Engl 49(22): 3830-3833.
- Shang X, Marchioni F, Evelyn CR, Sipes N, Zhou X, et al. (2013) Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc Natl Acad Sci U S A110(8): 3155-3160.
- Recchi C, Seabra MC (2012) Novel functions for Rab GTPases in multiple aspects of tumour progression. Biochem Soc Trans 40(6): 1398-1403.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.