Mini Review 
 Creative Commons, CC-BY
Creative Commons, CC-BY
A Synopsis of Different Plant LRR-RLKs Structures and Functionality
*Corresponding author: MHM Mubassir, Department of Mathematics and Natural Sciences, BRAC University, Bangladesh
Received: January 09, 2019; Published: January 29, 2019
DOI: 10.34297/AJBSR.2019.01.000517
Plant Defense Mechanism
As sessile organism plants are exposed to multiple environmental threats. Unlike animals, they lack mobile immune cells. Hence, to safeguard themselves from a range of pathogens, plants come by a multi-tiered innate immune system [1,2]. The first line of defense mechanism is the recognition of the pathogen by detecting pathogen-associated molecular patterns (PAMPs) or microbial-associated molecular patterns (MAMPs) which are either extracellularly exposed or secreted extremely conserved pathogen- derived molecules [1,3]. This recognition is achieved by a cognate plant cell-surface pattern-recognition receptor (PRR). Once the invading pathogen’s microbial signature ‘PAMP’ is recognized by the PRR, the plant cell initiates signaling cascades via the mitogen- activated protein kinase (MAPK). This first line of the plant defense mechanism is referred to as pattern-triggered immunity (PTI) [4- 8].
Pattern Recognition Receptors (PRRs)
PRRs are grouped into receptor-like kinase (RLK) and receptor-like protein (RLP). RLKs contain an extracellular domain, transmembrane domain, and intracellular kinase domain while RLPs lack kinase domain [6,7,9-11]. The number of RLKs and RLPs are much higher in plants compared to animals [12]. In Arabidopsis plant, around 410 RLKs and 170 RLPs have found so far, which is around 640 and 90 for rice consecutively [12-14]. Again, the number of RLKs is much higher than the number of RLPs. In the success of terrestrial plants, these high number of presence of RLKs and RLPs might have played a crucial role [15].
Based on the extracellular domain (ECD), RLKs and RLPs are again categorized into several subfamilies which include epidermal growth factor-like (EGF) domains, lysine motif (LysM) and leucine- rich repeat (LRR) [16]. Among these subfamilies, leucine-rich repeat like kinase (LRR-RLKs) is the largest family in plant [15]. With over 200 members of LRR-RLKs in Arabidopsis, they play a great role in plant development and defense processes which includes symbiosis, wounding response, hormone perception, stem cell maintenance and cell proliferation [17].
Structure of Leucine-Rich Repeat Receptor-Like Kinase (LRR-RLK)
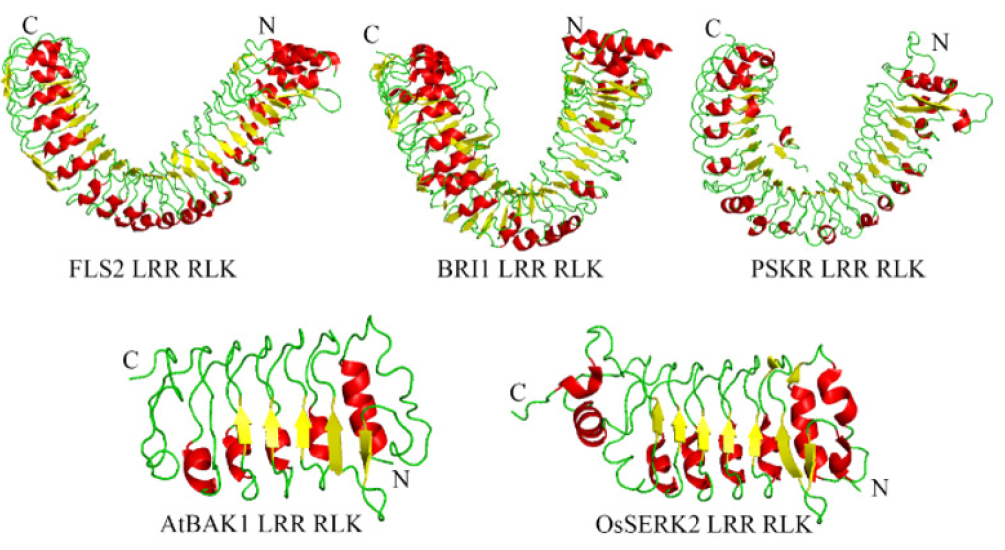
Plant LRR-RLKs are composed of a very specific consensus sequence LxxLxxLxLxxNxLSGxIPxxLGx, while x symbolizes non- conserved residues. This unique 24 amino acid sequence forms the beta sheet or beta turn which acts as protein-protein interaction surface. This unique motif forms a helical horse-shoe like structure in plant [18-21]. According to the recent studies, not all LRR-RLK domains composed of tandem arrays of multiple LRRs form horse- shoe like structure. Instead, some of them like plant brassinosteroid (BR) LRR form a right-handed superhelical structure which is different from solenoid conformation [19,22,23]. This unique GxIP sequence which form the helical structure is common in case if receptor-like protein kinase 2 (RPK2) [24], flagellin-insensitive 2 (FLS2) [25], BRI1-like receptor kinase 1 (BRL1) [26]. There is a notable difference in the size of LRR-RLK. Recent crystallographic structures reveal that this size varies from 21-19 LRRs. Recent studies showed that around 20-25 LRRs form this superhelical structure where inner surface and lateral side of the structure actively involves in binding with other proteins [25,27]. PGIP has 10 LRRs, SERK1 has 5 LRRs, HAESA has 21 LRRs, TDR/PXY has 22 LRRs, FLS2 has 29 LRRs, BRI1 has 25 LRRs, PSKR has 21 LRRs, RPK2 has 22 LRRs and TMK1 has 13 LRRs [28]. Small sized LRR- RLK is observed in AtBAK1 and OsSerk2 (Figure 1) which acts as a co-receptor for activation of PTI by PRR FLS2 and Xa21 respectively [25,29].
Island domain in LRR-RLK
In LRRs, LRR-RLK stands between 1st to 322nd LRRs where there can be a spacer region at 4th to 5th LRR from the C terminal end of 30 to 70 amino acids known as island domain (ID) [9]. The ID is not likely for all the LRR-RLK and its sequence is not conserved between species. These ID can interact with the PAMPs like BL with BRI1 [30] and PSK with PSKR [31,32] (Figure 2). Also, 2 ID were observed in the crystal structure of RPK2 which creates scaffold where the ligand binds [24]. This suggests the function of island domain as binding of LRR ectodomain with small ligands [28].
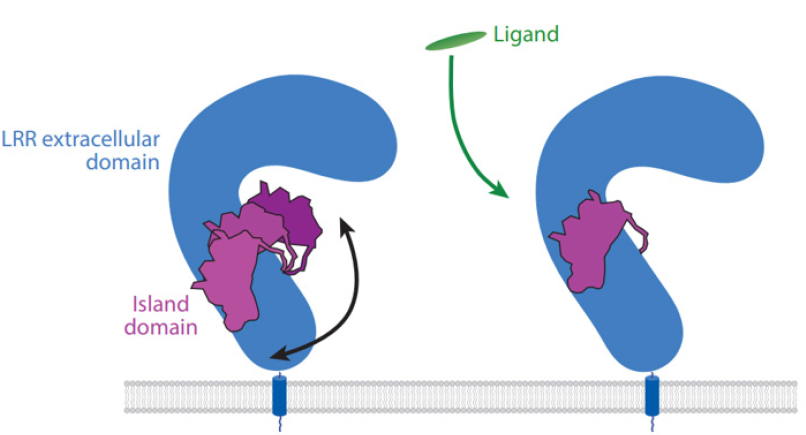
Figure 2: Island domain in BRI1 (left) and PSKR (right) [28].
Cys-pairs in LRR-RLK
Again, Cys-pairs which are placed between last LRR and transmembrane domain or near the start codon before LRR is another interesting extracellular domain in LRR-RLK [33]. Although the function of the Cys-pair is not clear, the mutation of it showed a significant decrease in FLS2 activity while BRI1 and CLAVATA2 showed no effect [34]. Although Cys-Pairs in LRR-RLK is assumed to contribute towards trafficking, folding and binding, no strict decision can be drawn regarding the importance of it.
Conclusion
The above discussion shows, in spite of the fact that most of the LRR-RLKs share a common shape, a significant difference is observed in the case of other factors such as diameter, LRR numbers, the presence of the island domain and cys-pairs. All this play a pivotal role in different binding mechanism in PTI complex at the early stage of the plant defense mechanism. It is needless to say, to obtain a precise idea about the detail function and interaction of different domains, further investigation needs to be done on LRRRLK conserved protein structures, motif compositions, and gene structures.
References
- Dodds PN, JP Rathjen (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nature Reviews Genetics 11(8): 539-548.
- Mott GA, Middleton MA, Desveaux, Guttman DS (2014) Peptides and small molecules of the plant-pathogen apoplastic arena. Frontiers in plant science (5): 677.
- Jones JD, JL Dangl (2006) The plant immune system. Nature 444(7117): 323-329.
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, et al. (1995) A receptor kinase- like protein encoded by the rice disease resistance gene, Xa21. Science 270(5243): 1804-1806.
- Medzhitov R, CA Janeway (1997) Innate immunity: the virtues of a nonclonal system of recognition. Cell 91(3): 295-298.
- Gómez Gómez L, T Boller (2000) FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Molecular cell 5(6): 1003-1011.
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, et al. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium- mediated transformation. Cell 125(4): 749-760.
- Ronald PC, B Beutler (2010) Plant and animal sensors of conserved microbial signatures. Science 330(6007): 1061-1064.
- Li J, J Chory (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90(5): 929-938.
- Gust AA, G Felix (2014) Receptor like proteins associate with SOBIR1- type of adaptors to form bimolecular receptor kinases. Current Opinion in Plant Biology 21: 104-111.
- Liebrand TW, HA van den Burg, MH Joosten (2014) Two for all: receptor- associated kinases SOBIR1 and BAK1. Trends in plant science 19(2): 123-132.
- Shiu SH, AB Bleecker (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci 98(19): 10763-10768.
- Fritz Laylin LK, Nandini Krishnamurthy, Mahmut Tör, Kimmen V Sjölander, Jonathan D.G. Jones (2005) Phylogenomic analysis of the receptor- like proteins of rice and Arabidopsis. Plant physiology 138(2): 611-623.
- Shiu SH, Wojciech M Karlowski, Runsun Pan, Yun-Huei Tzeng, Klaus F. X. Mayer, et al (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. The plant cell 16(5): 1220-1234.
- Li L, Yufei Yu, Zhaoyang Zhou, Jian-Min Zhou (2016) Plant pattern-recognition receptors controlling innate immunity. Science China Life Sciences 59(9): 878-888.
- Macho AP, C Zipfel (2014) Plant PRRs and the activation of innate immune signaling. Molecular cell 54(2): 263-272.
- Kobe B, AV Kajava (2001) The leucine-rich repeat as a protein recognition motif. Current opinion in structural biology 11(6): 725-732.
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, et al. (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474(7352): 467-471.
- She J, Zhifu Han, Tae-Wuk Kim, Jinjing Wang, Wei Cheng, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472-476.
- Di Matteo A, Federici L, Mattei B, Salvi G, Johnson KA, et al. (2003) The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine- rich repeat protein involved in plant defense. Proceedings of the National Academy of Sciences 100(17): 10124-10128.
- Kobe B, J Deisenhofer (1993) Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature 366(6457): 751- 756.
- Everitt B, T Hothorn (2011) An introduction to applied multivariate analysis with R: Springer Science & Business Media.
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, et al. (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275(5308): 1943-1947.
- Song W, Han Z, Sun Y, Chai (2014) Crystal structure of a plant leucine rich repeat protein with two island domains. Science China Life Sciences 57(1): 137-144.
- Sun Y, Li L, Macho AP, Han Z, Hu Z, et al. (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342(6158): 624-628.
- She J, Han Z, Zhou B, Chai, (2013) Structural basis for differential recognition of brassinolide by its receptors. Protein & cell 4(6): 475-482.
- Sun Y, Zhifu Han, Jiao Tang, Zehan Hu, Chengliang Chai, et al. (2013) Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell research 23(11): 1326-1329.
- Hohmann U, K Lau, M Hothorn (2017) The structural basis of ligand perception and signal activation by receptor kinases. Annual review of plant biology 68: 109-137.
- Caddell DF, Tong Wei, Sweta Sharma, Man Ho Oh, Chang-Jin Park, et al. (2018) Four tyrosine residues of the rice immune receptor XA21 are not required for interaction with the co-receptor OsSERK2 or resistance to Xanthomonas oryzae pv. oryzae. Peer J 6: e6074.
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, et al. (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433(7022): 167-171.
- Zhang Z, BP Thomma (2013) Structure‐function aspects of extracellular leucine‐rich repeat‐containing cell surface receptors in plants. Journal of integrative plant biology 55(12): 1212-1223.
- Wang J, Hongju Li, Zhifu Han, Heqiao Zhang, Tong Wang, et al. (2015) Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525(7568): 265-268.
- Diévart A, SE Clark (2003) Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Current opinion in plant biology 6(5): 507-516.
- Sun W, Cao Y, Jansen Labby K, Bittel P, Boller T, et al. (2012) Probing the Arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. The plant cell 24(3): 1096-1113.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.