Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
A Comprehensive Overview of Deep Learning Techniques for Retinal Vessel Segmentation
*Corresponding author: Adamopoulou M, Department of Health Physics and Computational Intelligence, School of Health Rehabilitation Sciences, University of Patras, Greece.
Received: July 29, 2023; Published: August 08, 2023
DOI: 10.34297/AJBSR.2023.19.002631
Abstract
The application of new techniques in eye imaging has revolutionized the process by increasing accuracy and efficiency levels considerably. An essential aspect of analyzing ocular images is the correct segmentation of retinal vessels. Deep learning algorithms represent an effective solution giving high accuracy rates for this task. In this article readers can find an extensive overview of several novel approaches built specifically to address this issue-among them being a groundbreaking deep learning architecture that applies multi-scale/multi-level Convolutional Neural Networks (CNNs) to obtain a comprehensive hierarchical representation of retinal vessels. Furthermore, advanced approaches are also discussed on detecting retinal vessels from Fluorescein Angiography (FA) images - with notable examples being one that combines cross modality transfer with human in the loop learning helping reduce manual labelling efforts significantly while enhancing both accuracy and performance metrics simultaneously. These stateof- the-art methods unravel critical insights into how deep learning methods could improve retinal vessel segmentation. The impact they could have on diagnosing and treating various eye related illnesses is also noteworthy.
Keywords: Eye imaging, Deep learning, Retinal vessel segmentation, Deep vessel architecture
Introduction
The progress made in eye imaging technology is commendable especially with regards to deep learning techniques employed for ocular image analysis. The benefits are evident as these methods hold great promise for improving accuracy and efficiency during retinal vessel segmentation-an important aspect of eye imaging practice. In this article we delve into this topic deeply by providing an overview of several innovative approaches being developed to tackle this issue. A standout technique among them includes “Deep Vessel” architecture introduced by Huazhu Fu, et al.,[1]. In their inventive method they utilized a multi-level Convolutional Neural Network (CNN) working at different scales to extract detailed hierarchical representations of retinal vessels while simulating long range interactions between pixels using Conditional Random Field (CRF). As expected with such sophisticated design approaches these components blend smoothly into a single cohesive deep network resulting in impressive outcomes during testing trials. Other notable cutting-edge methods highlighted in this article are approaches by Li Ding, et al., [2] for identifying retinal vessels in Fluorescein Angiography (FA) images. Efficiently segmenting retinal vessels has long been an area with many challenges, but recent developments in deep learning are already transforming this field. A fascinating article delves into an innovative combined approach using cross-modality transfer along with human-in-the-loop learning to minimize manual labour while boosting accuracy and overall performance levels significantly. These techniques offer tremendous potential for advancing the diagnosis as well as treatment of eye disorders through sophisticated ocular imaging techniques.
State of the Art
In the realm of eye imaging, recent breakthroughs have high lighted the application of deep learning methods for examining ocular images. Retinal vessel segmentation is a crucial aspect of ocular imaging, boasting a multitude of uses. A trailblazing team of researchers, spearheaded by Huazhu Fu, et al., [1], tackled this issue by viewing it as a boundary detection task and introducing a groundbreaking deep learning architecture known as the “Deep Vessel.”
This inventive method revolves around two primary concepts. Firstly, a multi-scale and multi-level Convolutional Neural Network (CNN) was employed, incorporating a side-output layer that facilitated the extraction of a comprehensive hierarchical representation of retinal vessels. This approach allowed the network to detect subtle details and features that might otherwise go unnoticed. Secondly, the researchers simulated long-range interactions between pixels using a Conditional Random Field (CRF). By merging these two layers, the “Deep Vessel” was born, forming a cohesive deep network that smoothly integrates the CNN and CRF components.
In a series of experiments carried out by a group of researchers, the “Deep Vessel” system proved to be highly effective. Its top-ofthe- line performance in segmenting retinal blood vessels within the DRIVE, STARE, and CHASE DB1 datasets was evident. These discoveries underscore the vital importance of deep learning methodologies in examining ocular imagery and set the stage for future breakthroughs within the field of eye imaging. The “Deep Vessel” framework, devised by Huazhu Fu’s team, offers a potential solution to the challenge of retinal vessel segmentation and could greatly impact the diagnosis and treatment of a variety of eye-related illnesses.
The implementation of deep learning techniques within the realm of ocular imaging has garnered considerable interest in recent times. Fu and his fellow researchers zeroed in on the issue of segmenting blood vessels within eye images, assessing and critiquing established methods like machine learning, adaptive models, and blood vessel tracking techniques. Ultimately, the research team introduced an innovative deep learning structure -the Deep Vessel- which achieved unparalleled success in the area of retinal blood vessel segmentation.
The trailblazing group headed by Li Ding, et al., [2] has created a cutting-edge approach for identifying retinal vessels in Fluorescein Angiography (FA) images through the use of Deep Neural Networks (DNNs). This revolutionary process considerably lessens the manual labour needed for annotating ground truth data, which is crucial for developing precise and efficient DNNs for retinal vessel identification. The process is composed of two primary components: cross-modality transfer and human-in-the-loop learning. The process of cross-modality transfer is facilitated through the simultaneous acquisition of Colour Fundus (CF) and fundus FA images. Initially, a pretrained neural network extracts binary vessel map from the CF images. These maps are then aligned and transferred to FA images using a sturdy parametric chamfer alignment applied to an unsupervised preliminary identification of FA vessels. This stage progressively enhances the accuracy of ground truth labelling by integrating the transferred vessels as the initial ground truth labels.
The human-in-the-loop learning segment of the process is an ongoing interplay between deep learning and manual annotation, which improves the quality of ground truth labelling, lessens the manual labour required for annotation, and facilitates expert involvement in the procedure. The authors of this innovative process examine several essential aspects of the methodology and demonstrate its performance on three distinct datasets. The findings reveal that the process surpasses existing FA vessel detection techniques and provides an accurate solution for identifying retinal vessels in FA images.
Moreover, the team has developed a new publicly available dataset, RECOVERY-FA19, consisting of high-resolution ultra-widefield images and precisely labelled ground truth binary vessel maps. This dataset serves as a valuable asset for researchers in the field and fosters the advancement of deep learning algorithms for retinal vessel identification in FA images.
In summary, the process proposed by Ding, et al., [2] signifies a considerable breakthrough in medical imaging and holds the potential to transform how retinal vessels are detected in FA images. The combination of cross-modality transfer and human-in-theloop learning substantially reduces manual labelling efforts while enhancing accuracy and performance. This work has far-reaching consequences for diagnosing and treating eye conditions and offers a promising solution for identifying retinal vessels in FA images.
Zengqiang Yan, et al., [3] explored the use of deep learning techniques for retinal blood vessel segmentation, which is crucial for diagnosing and treating eye-related conditions. Conventional deep learning approaches using pixel-wise losses struggle to accurately segment narrow vessels due to the uneven proportion of narrow and wide vessels’ thickness in fundus images. The researchers proposed a novel segment-level loss that focuses on maintaining the consistency of narrow vessel thickness during training. Combining segment-level and pixel-level losses, the joint-loss framework balances the weights assigned to wide and narrow vessels, enhancing vessel segmentation. This approach could significantly influence the diagnosis and treatment of eye-related disorders by providing a more accurate and efficient method for retinal vessel segmentation.
Wang, et al., [4] developed a deep learning framework based on the traditional Convolutional Neural Network (CNN) model, U-net, to precisely pinpoint the Optic Disc (OD) in colour fundus images, which is crucial for detecting and diagnosing retinal diseases. The two-phase framework consists of training the U-net model with colour fundus images and monochromatic vessel density maps and using an overlap technique to combine the segmentation results. The proposed framework outperformed the standalone U-net model, achieving high accuracy in OD segmentation. While the study shows the potential of this deep learning framework, further research is needed to confirm its clinical applicability and effectiveness. Nonetheless, the results offer a promising outlook for future advancements in deep learning for retinal disease diagnosis.
Pan Xiuqin, et al., [5] addressed the challenges of existing retinal vessel segmentation methods by developing a technique using an enhanced U-Net deep learning model. The team improved retinal image quality and incorporated a residual module into the network design to address the insufficient depth of traditional U-Net models. The approach was evaluated using the DRIVE dataset and achieved impressive results in segmenting retinal blood vessels, even with pathological data. The study highlights the advantages of incorporating residual modules into U-Net models for retinal vessel segmentation, but further research is needed to validate its clinical utility and effectiveness. The results provide promising evidence for future advancements in deep learning for retinal disease diagnosis.
Wu, et al., [6] investigation in 2019 focused on combining residual network architecture with DenseNet to improve learning retinal blood vessel morphological structures but faced challenges due to the Dense Block framework’s memory requirements and computational complexities. Zhang, et al., [7] mf-fdog matching filtering approach and Vlachos and Dermatas, et al., [8] iterative line tracking program have their advantages and drawbacks. Model-based methods and Espona, et al., [9] serpentine model, combined with morphological operations, offer alternative approaches. Toufique Soomro, et al., [10] examined over 80 literature pieces on retinal vessel detection and found only 17 papers addressing segmentation using deep learning techniques. The study provides an in-depth analysis of various deep learning strategies, such as CNNs, RNNs, and GANs, highlighting their strengths and limitations. The authors suggest future advancements in retinal image examination by developing innovative deep learning methods and combining existing techniques for improved precision and efficacy. Erick Rodrigues, et al., [11] introduced a method for retinal vessel segmentation that combines region growing and machine learning techniques. The unique feature extraction approach uses grey level and vessel connectivity characteristics for better information dissemination during classification. The method outperformed 25 of the 26 compared techniques on the DRIVE dataset and achieved high accuracy scores on STARE, CHASE-DB, VAMPIRE FA, IOSTAR SLO, and RC-SLO datasets. The approach has the potential to improve accuracy and efficiency in retinal vessel segmentation.
Maninis, et al., [12] introduced Deep Retinal Image Understanding (DRIU), a framework that uses Convolutional Neural Networks (CNNs) to address retinal vessel and optic disc segmentation. The DRIU system surpassed human annotators in all instances and showed consistency with the gold standard. This work demonstrates the potential for a unified approach to retinal vessel and optic disc segmentation, with implications for the development of advanced diagnostic tools for ocular diseases. The findings suggest that DRIU could reshape retinal image analysis and pave the way for future advancements in the field (Table 1).
Applications of AI in Detecting Abnormalities and Diseases in Ophthalmic Imaging
Manual segmentation of retinal vessels is an intricate process known for demanding both knowledge and finesse from medical professionals who engage with it daily. While these experts possess considerable expertise on the matter along with years of experience under their belt; nevertheless; complications arise due to potential human errors that cause inaccuracies during manual processing consistently-Adding significance towards automating solutions for retinal vessel segmentation within healthcare circles recently accomplished by researching groups applying machine learning towards resolving inconsistencies caused by human error during manual processing with cutting-edge technology that improves accuracy significantly being developed as we speak.
By employing a semi-supervised clustering method, the researchers were able to identify similar instances from various databases, enabling the algorithm to effectively train on both labelled and unlabelled data. To their delight, they found that images from different classes contributed to better performance compared to those from the same class. The trained network was then tested on three different datasets, DRIVE, STARE, and HRF, and achieved an accuracy rate of 97%, surpassing manual segmentation by human raters. Upon conducting cross validation tests, it became evident that the trained network maintained a high level of accuracy at a rate between 80% to 83%. The application of machine learning methods for retinal vessel segmentation presents significant opportunities for transforming diagnosis within medicine. A more efficient and reliable tool that reduces the likelihood of human error can be achieved.
The proposed framework’s remarkable accuracy rate and ability to utilize both labelled and unlabelled data make it a promising solution for retinal vessel segmentation in the medical realm [13]. In their research, Theodoros Pissas, et al., [14] delve into the intricacies of retinal blood vessel segmentation in Optical Coherence Tomography Angiography (OCT-A) images of the human retina, with the goal of enhancing image-guided therapy administration during vitreoretinal surgery. The authors underscore the distinct advantages of OCT-A scans, as they offer a high-resolution perspective of the macular blood vessels, an essential element for achieving favourable surgical results.
The authors present a novel method to segment the Superficial Vascular Plexus using Convolutional Neural Networks (CNNs) in 2D Maximum Intensity Projections (MIP) of OCT-A images. The CNNs are designed to iteratively improve the quality of vessel segmentations, and the method is evaluated using a sample of 50 subjects, including those with structural macular abnormalities and healthy individuals.
The results reveal that the proposed approach outperforms other network- and graph-based methods and demonstrates its generalizability to three-dimensional segmentation and OCT-A scans with narrower fields of view. In conclusion, the authors emphasize the importance of precise and automated extraction of detailed vessel maps in retinal surgery and how their method paves the way for the development of more advanced techniques for guiding regenerative therapy delivery.
The authors’ work highlights the significance of utilizing advanced computational techniques to optimize surgical planning and improve patient outcomes in retinal surgery. The research by Hyeongsuk Ryu, et al., [15] introduces a novel era of medical image analysis, owing to their pioneering Direct Net architecture. This structure harnesses the power of deep learning to precisely detect retinal vessels, surpassing patch based CNNs with its exceptional results. The Direct Net consists of blocks arranged in a pyramidal fashion, each comprising a series of convolutional layers that process and combine data as they ascend. This recurrent configuration and the use of a large kernel eliminate the need for up- or down sampling layers, resulting in a direct mapping of pixel inputs to segmentation outputs.
The performance of the Direct Net is remarkable, with accuracy scores reaching 0.9538, outperforming the patch-based CNN’s score of 0.9327. Its sensitivity score of 0.7851 is superior to the patch-based approach’s score of 0.7346, and the specificity score of 0.9782 is higher than the patch-based CNN’s score of 0.9730. The Direct Net also boasts a precision score of 0.8458, surpassing the patch-based CNN’s score of 0.7987. Besides its impressive performance, the Direct Net architecture has significantly reduced training and testing times. The training time has been shortened from 8 hours to merely 1 hour on a standard dataset, while the testing time per image has been reduced from 1 hour for the patch-based method to a mere 6 seconds for the Direct Net. These improvements make this architecture a valuable resource for medical professionals handling large volumes of data who require rapid and accurate results.
In conclusion, the Direct Net architecture by Hyeongsuk Ryu, et al., [15] represents a groundbreaking accomplishment in the realm of medical image analysis, with its recurrent structure, large kernel, and direct mapping capabilities yielding improved accuracy, sensitivity, specificity, and precision. The reduction in training and testing times renders it an attractive option for medical professionals working with extensive amounts of data.
Exploring Other Imaging Modalities in Ophthalmic Imaging
In the domain of medical imaging, Pavle Prentasic, et al., [16] investigated blood vessel segmentation utilizing deep learning networks. They applied Convolutional Neural Networks (CNNs) to distinguish between vessels and non-vessels in Optical Coherence Tomography Angiography (OCT-A) images. The CNNs were composed of a combination of convolutional and max pooling layers that transformed raw pixel intensities into feature vectors, which were subsequently classified by fully connected layers.
In their groundbreaking research, Pavle Prentasic, et al., [16] and their innovative team sought to evaluate the effectiveness of deep learning algorithms in segmenting foveal microvasculature within OCT-A images. By employing deep convolutional neural networks, they managed to achieve automated segmentation, classifying each pixel as either vessel or non-vessel.
The researchers trained the neural networks using a balanced set of original OCT-A images and their corresponding manual segmentations, ensuring an equal distribution of vessel and non-vessel pixels in each instance. Cross-validation test results demonstrated promise, as the trained network achieved accuracy percentages between 80% and 83%, on par with the accuracy of manual segmentation performed by human raters. However, the study also highlighted significant variability in manual segmentation, both intra- and inter-rater, indicating that the trained network might even surpass the performance of a newly trained human rater.
The automated segmentation process took a mere 2 minutes, in stark contrast to the 20 to 25 minutes required for manual segmentation by human raters. This remarkable speed offers a valuable tool with the potential to expedite medical image analysis in clinical settings and deliver faster results to healthcare professionals. In summary, this study emphasizes the vast potential of deep learning networks in transforming medical image analysis procedures and facilitating more efficient outcomes.
Classification Methods
Deep learning algorithms have gained immense attention in the world of ophthalmic imaging, showcasing their ability to delve into eye images and reveal previously unattainable insights. Most research in this area focuses on harnessing the power of basic Convolutional Neural Networks (CNNs) for analyzing Color Fundus Imaging (CFI) data.
The scope of deep learning algorithms in ophthalmic imaging is vast, encompassing tasks such as segmenting eye anatomy, detecting retinal abnormalities, diagnosing eye diseases, and even assessing image quality. A key demonstration of the impact of these algorithms can be seen in the 2015 Kaggle competition, where over 35,000 colour fundus images were utilized to train deep learning models to estimate diabetic retinopathy severity in a set of 53,000 test images.
Numerous participating teams employed deep learning algorithms, with four of them even surpassing human experts by using end-to-end CNNs. In another study by Gulshan, et al., [17], the performance of a Google Inception v3 network in identifying diabetic retinopathy was found to be on par with a group of seven certified ophthalmologists. These findings underscore the tremendous potential of deep learning algorithms in delivering faster, more efficient, and precise results in ophthalmic imaging.
The realm of retinal vessel segmentation in colour fundus, Fluorescein Angiography (FA), and Scanning Laser Ophthalmoscopy (SLO) images has undergone a significant transformation due to innovative techniques. This overview delves into the recent advancements in the field, emphasizing both supervised and unsupervised machine learning approaches. However, to provide a comprehensive understanding, it also covers the latest non-machine learning techniques, which are organized into four categories: morphological image processing, vessel tracing/tracking, multi-scale, and other methods [18].
Morphological image processing uses mathematical morphological operations to process images and isolate features. Techniques like morphological gradient extract vessel-like structures by removing background pixels. Vessel tracing/tracking follows vessel paths to trace retinal vessels, with accurate tracing and avoiding false detections as key challenges.
The multi-scale category processes images and extracts features at multiple scales, crucial for retinal vessel segmentation, where vessels have varying sizes and shapes. A single scale makes accurate segmentation difficult. The “other methods” category covers techniques that don’t fit the previous categories, including phase-based, deep learning-based, and region-growing methods. These techniques are also discussed in-depth, emphasizing key features and performance.
In conclusion, this comprehensive overview presents a detailed look at recent advances in retinal vessel segmentation, centred on various techniques and their performance. An appendix table summarizing each paper makes comparing and evaluating different techniques easier.
Machine Learning Methods
When it comes to machine learning techniques one can classify them into two categories based on their approach: supervised or unsupervised techniques. The former includes Bayesian approaches, SVMs, ANNs, random forest algorithms, AdaBoost decision trees along with deep learning systems that rely on ground truth labels for training classification models. These methods are generally more accurate, but their success depends heavily on the quality and quantity of labelled datasets [19,20]. Unsupervised methods such as GMMs, FCMs or k means clustering approach data analysis without relying on training labels thus enabling them to reveal hidden patterns which may otherwise be missed by supervised methodologies. Nevertheless, they may not be as precise as supervised techniques resulting in lower accuracy rates. Therefore, it is crucial for researchers to focus both on the application requirements along with labelling availability before deciding which approach would best suit their project.
Deep Learning
Deep learning models, especially CNNs, excel at classification and categorization tasks. They have been used for vessel segmentation, either independently or combined with other techniques like random forests [12]. Other approaches include using CNNs with CRFs [21]. Modified architectures with specialized layers have been proposed for combined vessel-optic disk detection [22], and CRF layers have improved lesion segmentation [23]. Dense CRFs models shorten training time [24]. Dasgupta and Singh (2017) segmented thin vessels, while FCNs showed better results than networks with few fully connected layers [25]. Multi-scale analysis and GANs have also been explored to improve segmentation performance [25,26]. Overall, deep learning techniques continue to advance vessel segmentation processes.
Other Machine Learning Methods
Supervised Methods
Backpropagation NNs are popular in retinal vessel segmentation, with various strategies to improve performance, such as incorporating texture [27], colour [28], intensity [29,30], and moment invariants [29]. Lattice NNs have been used to enhance convergence [29]. Cross-modality learning, and deep architectures have also been applied for segmenting vessels in noisy images [31]. Thin vessels have been segmented using feature descriptors like LBP and shape features [31].
Combining NNs with LDA involves integrating Gaussian, Wavelet, and Gabor filters with intensity, Hessian-based, and SIFT features for vessel enhancement and segmentation [32-34]. Annunziata and Trucco [35] presented an approach using SCIRD-TS filters for segmenting thin vessels. Ensemble learning algorithms like bagging and boosting have shown effectiveness in vessel segmentation [19,30,36,37], particularly the AdaBoost algorithm in capturing vessel information from normal and pathological pediatric retinal images [30].
Fuzzy inference combined with multi-scale LBP, Gaussian, and directional features has been used for vessel segmentation [38,39]. Dictionary learning and sparse representations have also shown improvements in vessel classifiably [40]. Techniques such as B-COSFIRE and Frangi filters have been used to enhance vessels [36], while ensemble features with divergence vector field have boosted pathological image performance [41,42].
SVM, when combined with fully connected conditional random fields, has improved segmentation results in computer vision tasks [43,44]. SVM has been used with k-means clustering, binary Hausdor symmetry measure, Gabor filters, B-COSFIRE filters, and multi-fractal features to achieve lesion-resistant vessel segmentation [45, 46-50]. Jebaseeli, et al., (2016). Random forest classifiers, combined with visual attention modelling, have been applied to address challenges in images with closely situated parallel vessels, CVR, low-contrast thin vessels, lesions, and non-uniform illumination [51,52]. These methods demonstrate the adaptability of SVM and its ability to tackle various segmentation challenges when combined with other techniques (Table 2).
Unsupervised Methods
Unsupervised vessel segmentation in medical imaging uses mathematical algorithms to classify pixels as vessels or non-vessels without human involvement. The Gaussian Mixture Model-Expectation Maximization (GMM-EM) is a popular algorithm for maximum- likelihood vessel/non-vessel classification, enhancing vessels through high-pass filtering and top-hat transformation [53]. GMMGray voting has also improved thin vessel detection and reduced vessel fragmentation Dai, et al., (2015). Fuzzy C-Means (FCM) clustering amplifies thin vessels and removes blobs from fundus images using filters like Frangi’s, Matched, and Gabor wavelets [54,55]. Evolutionary algorithms, such as Bee Colony Optimization, have been used to identify vessel clusters and distinguish vessels from background pixels [56,57]. Overall, unsupervised vessel segmentation techniques offer improved results and do not rely on human input, providing enhanced outcomes in medical image analysis.
Matched Filtering Methods
The Matched Filtering (MF) method is an established technique in image processing that compares retinal images with pre-designed kernel models, imitating the intensity profiles of vessels [58-61]. To improve visibility of slender, low-contrast vessels in images with lesions and bright blobs, various strategies have been employed, such as curvelet transform, Laplacian of Gaussian filter [62-64], two-dimensional Gabor filtering, multi-scale line detection, anisotropic diffusion, and B-COSFIRE filtering [60,61,65,66]. Support Vector Machines (SVM) can be used for segmentation by merging contrast and diffusion maps [65]. Spline fitting, length, and adaptive filtering can help refine segmented vessels and minimize artifacts [61,67]. Parameter adjustments for Gabor filters can be done using optimization algorithms like Particle Swarm Optimization (PSO) and Imperialism Competitive Algorithm (ICA) [59,68,69]. Additionally, MF has been studied within portable FPGA-based hardware architectures [70,71]. In summary, numerous techniques are available to enhance and distinguish slender, low-contrast vessels in challenging images, and ongoing advancements in image processing promise new, more effective methods.
Morphological Image Processing Methods
Mathematical morphology is a potent technique in digital image processing, crucial for identifying boundaries, skeletons, and convex hulls [72]. Gonzalez and Woods (1988). In this field, morphological operators are used to carry out a variety of tasks on binary images, including connecting disjointed regions, filling gaps, and shrinking objects, with dilation and erosion being the most commonly used operators Serra, et al., (1979).
A primary application of mathematical morphology in digital image processing involves improving vessel visibility, often using the top-hat transformation to estimate image background through morphological opening operations [19,73]. Vessel enhancement in images can be accomplished through methods such as bit plane slicing, multi-directional morphological top-hat transform, and the H-maxima transform [74].
Mathematical morphology in digital image processing offers several advantages over alternative techniques. Morphological operators are non-linear and non-iterative, rendering them ideal for real-time applications due to their computational efficiency. Furthermore, morphological operators facilitate the preservation of geometrical information in the image, making them invaluable in applications where geometrical details are essential.
In summary, mathematical morphology is a foundational aspect of digital image processing, with a wide range of applications, from vessel enhancement to the detection of boundaries, skeletons, and convex hulls. The non-linear and non-iterative nature of morphological operators renders them a computationally efficient solution, well-suited for real-time applications. As computational power and image processing algorithms continue to advance, the utilization of mathematical morphology in digital image processing is expected to grow significantly.
The complex field of vessel centerline segmentation, a crucial topic within the medical imaging domain, has been the subject of extensive investigation and development. A notable milestone includes [75] innovative method, which combined FoDoG (First Order Derivative of Gaussian) and iterative region growing to identify vessel centerlines, culminating in a binary map via multilevel thresholding [75]. The efficacy of separating vasculature and pathological lesions using the Morlet Wavelet Transform (MWT) and Morphological Component Analysis (MCA) has also been demonstrated [76].
A combination of different techniques, including FoDoG, adaptive thresholding, and morphology-based global thresholding, has been employed to accurately detect thin vessels [76,77]. In the presence of occlusions, the Hidden Markov Model (HMM) has demonstrated its effectiveness in tracking vessels and segmenting both thin and thick vessels [78]. By using mean-C thresholding and morphological cleaning, Dash and Bhoi [79] have improved segmentation accuracy by eliminating disconnected areas.
In conclusion, vessel centerline segmentation remains an active area of research, with numerous methods being developed to achieve precision in identifying vessel centerlines and differentiating them from pathological lesions. The integration of various techniques and the use of models like HMM have proven invaluable in ensuring accuracy, even in the presence of occlusions.
Vessel Tracing and Tracking Methods
The complexities of vessel tracing and tracking require the use of seed points, chosen from the borders or centerlines of the vessels, as a starting point. These seed points, based on local data, provide insights into vessel dimensions and the intricacies of their interconnectivity at complex crossover points and bifurcations. The intersection and crossing of vessels can be determined through a combination of particle filtering, Kalman filtering, and hysteresis thresholding [80,81].
A multi-scale, multi-orientation filtering approach, known as MF, has been employed to construct maps of vesselness, enhancing the visual representation of vessels [82,83]. Techniques such as minimum-cost matching, Dijkstra’s algorithm, and global graph optimization help maintain the continuity and connectedness of traced vessels. These methods have proven successful in segmenting low-contrast, tortuous vessels found in retinal photographs of premature infants [80,81,84].
The intricate task of distinguishing vessel segments as arteries or veins is adeptly managed by combining snakes, gradient orientations, and minimum pathways through the implementation of k-means clustering Vázquez, et al., (2013). Utilizing line and orientation detection across multiple scales assists in tracing vessel edges and centrelines in complex regions near vessel crossovers and bifurcations [85].
To amplify the visibility of fragile vessel segments, researchers have integrated the Firangi filter response [86,87] into their approach. This filter enhances thin and elongated structures in images, making it well-suited for vessel segmentation. Otsu’s thresholding and tensor colouring were used to complement this technique, producing binary maps of the targeted vessels [87]. These inventive methods have demonstrated their worth in examining and visualizing retinal vessels, with precise tracking of edges and centrelines being essential for diagnosing and treating eye conditions such as diabetic retinopathy, age-related macular degeneration, and glaucoma. Employing k-means clustering, gradient orientations, and minimum pathways facilitates the identification of vessel segments as arteries or veins, a critical step in addressing these diseases.
In conclusion, the accurate classification of vessel segments as arteries or veins, along with precise tracking of edges and centrelines, is essential for effective retinal vessel analysis. The combination of snakes, gradient orientations, minimum pathways, and k-means clustering provides a robust solution for these challenges. The Frangi filter response, Otsu’s thresholding, and tensor colouring are instrumental in enhancing the visualization of thin vessel segments that are often difficult to detect. These techniques have proven their efficacy in the diagnosis and treatment of eye diseases and will continue to be pivotal in retinal vessel analysis.
Artery/Vein Classification Methods
During the period examined, the number of research papers specifically targeting arteriole and venule (A/V) classification is significantly less than those focusing on vessel segmentation. Traditional Convolutional Neural Networks (CNNs) have been employed for the classification of arteries and veins, exhibiting success in segmenting delicate vessels even when occlusions are present [88].
Probability graph propagation has shown effectiveness in correcting misclassified pixels [89,90].
Furthermore, Fully Convolutional Network (FCN) segmentation of arteries and veins using the U-Net architecture has been documented [91,92]. Generative Adversarial Networks (GANs) with topological structure constraints and adversarial loss have displayed greater efficiency in learning the probability distribution of arteriovenous segmentation maps compared to other methods [93].
In the context of vessel classification and Arteriole-to-Venule Ratio (AVR) estimation, Linear Discriminant Analysis (LDA) has been applied alongside intensity, retinex normalization, and reflection properties of the vessels [94-96]. The AVR is a well-regarded metric in retinal biomarker research [91] and represents the ratio of the weighted average width of arterioles to that of venules in the area around the optic disc.
Bayes classifiers and graph cut techniques have been used by Pellegrini, et al., [97] to analyze Ultra-Wide Field of View (UWFoV) Scanning Laser Ophthalmoscopy (SLO) images. This highlights the diverse range of methodologies and techniques researchers have employed to investigate and improve upon A/V classification and vessel segmentation in retinal imaging.
The advancements in deep learning and various machine learning techniques have paved the way for more sophisticated approaches to A/V classification and vessel segmentation. These innovative methods are instrumental in enhancing the accuracy and efficiency of retinal image analysis, ultimately leading to improved diagnosis and treatment of retinal diseases. The continuous exploration and development of these techniques contribute to a better understanding of retinal biomarkers, ensuring more reliable and precise results in retinal biomarker research.
The classification of blood vessels in retinal images has been explored using a range of classification and clustering techniques that leverage various features. Support Vector Machines (SVM) have been employed by numerous researchers [98-100] Vijayakumar, et al., (2016) to analyze width, orientation, Gabor, intensity, and morphological features, as well as to execute feature selection through Random Forest (RF) and graph-theoretic frameworks with vessel tree network topology Estrada, et al., [101].
K-Nearest Neighbors (KNN) has been assessed using multiscale, colour, texture, and adaptive Local Binary Pattern (LBP) features by various investigators [41,102-104]. Additionally, k-means clustering with color features has been applied to classify arteries and veins within specific sections of fundus images. To compute the Arteriole-to-Venule Ratio (AVR), Fu, et al., [105] and Relan, et al., [106,107] have used these methods.
A more advanced approach involved Joint Boost, which utilizes vessel network topology Yan S, et al., [108]. Vessel tracing and optimal forest graph representations have been integrated into the widely recognized Singapore I Vessel Assessment (SIVA) system, which has been used in numerous clinical studies [109].
Furthermore, Eppenhof, et al., [110] have examined energy functions based on Markov Random Fields (MRFs), RF classifiers with Gabor wavelet and statistical histogram features, vessel key point detection, and metaheuristic graph search techniques. This approach has been further investigated by Srinidhi, et al., [111].
In summary, a variety of methods have been investigated in the context of retinal vessel classification, including deep learning, supervised and unsupervised techniques, vessel tracing and tracking, and multi-scale approaches. These diverse methodologies reflect the ongoing efforts of researchers to develop more accurate, efficient, and robust techniques for vessel classification in retinal images. As the field continues to evolve, the incorporation of advanced machine learning algorithms and feature extraction techniques promises to enhance the accuracy and reliability of retinal vessel classification, ultimately leading to improved diagnostic capabilities and more effective treatments for retinal diseases.
The detection and diagnosis of retinal diseases present significant challenges for medical practitioners, as these conditions can be severe, and results from image processing techniques may be inconsistent. To address this issue, a novel approach has been proposed that combines various stages for improved accuracy.
The initial phase, pre-processing, focuses on cleaning the images of noise and enhancing their quality, followed by defining a Region of Interest (ROI) to extract specific attributes and segmentation. Subsequently, a Deep Convolutional Neural Network (DCNN) is combined with other techniques like HOG, LBP, and SURF to extract features from the ROI, including textural properties, contrast, scaling, rotation, and translation [112].
A feature selection process is then executed to eliminate unimportant features and choose the most representative ones, which are then inputted into multi-class classifiers such as SVM, KNN, and RF for classification into either diseased or healthy classes. The proposed system leverages the power of deep learning to provide a more precise and trustworthy method for retinal disease detection. In summary, the proposed methodology incorporates multiple stages of image processing and feature extraction to achieve a more accurate diagnosis. The integration of deep learning and various techniques promises to enhance the reliability and precision of retinal disease detection (Table 3).
Comparative Results
From the presentation thus far, it becomes evident that drawing fair and consistent comparisons between the multitude of reported methods is far from a straightforward task. This section delves into the primary concerns surrounding this issue and provides suggestions on how to navigate these challenges. A crucial aspect of the broader field of medical image validation is the availability, design, and objectives of public datasets, which has been acknowledged as a critical component sparking substantial international discussions [113-116]. This debate encompasses a variety of international grand challenges that are characterized by well-established, yet often contrasting, performance metrics.
In this section, the authors examine two instances of retinal image analysis, although their primary focus is not on vessel detection and labelling. The first instance involves REFUGE at MICCAI, which is linked to the OMIA workshop on retinal image analysis that occurred on March 13th. REFUGE 2018 accumulated a set of 1,200 images, with 400 allocated for training purposes. These images were annotated by seven independent specialists in optic disc detection, glaucoma classification, and fovea localization. The second instance is the IDRID (Indian DR Image Dataset) challenge, conducted during ISBI 2018 (https://idrid.grand-challenge.org/). This challenge emphasizes DR identification and classification, as well as diabetic macular enema. Numerous other comparable challenges can be readily found online.
Based on a quantitative evaluation of the results from recent MICCAI Grand Challenges, Maier Hein, et al., [113] offer fascinating insights and observations. These grand challenges play a crucial role in progressing the field of medical image analysis by uniting researchers and experts from across the globe. They establish a platform for the creation and comparison of innovative techniques and algorithms, promoting collaboration and inventiveness. The shared datasets, evaluation metrics, and best practices contribute to advancements in medical imaging technology, ultimately benefiting both medical professionals and patients.
One key factor contributing to the difficulty in comparing different methods is the wide range of datasets and evaluation criteria used in these challenges. The quality and representativeness of the datasets play a significant role in determining the robustness and generalizability of the developed methods. Furthermore, the evaluation criteria must be carefully designed to ensure that they measure the performance of the methods in a fair and meaningful way. This may involve considering factors such as the complexity of the images, the presence of various confounding factors, and the specific clinical applications of the developed techniques.
Another challenge in comparing the methods is the often-proprietary nature of some datasets and algorithms, which can hinder the transparency and reproducibility of the results. This underscores the importance of open science practices, including the sharing of datasets, code, and detailed methodological descriptions, to enable a more rigorous and fair comparison of the different methods. In conclusion, the complexity of comparing the numerous methods reported in the field of medical image validation highlights the need for a more systematic and standardized approach. By addressing the challenges related to the availability, design, and purpose of public datasets, as well as the evaluation criteria used in international grand challenges, the research community can continue to make progress and drive advancements in medical imaging technology.
The majority of techniques documented for segmenting retinal vessels have been evaluated using the DRIVE database. Additional assessments have been carried out on databases such as STARE [117] CHASEDB1 [118], and HRF [119]. Moreover, numerous methods have been examined on other datasets, comprising both publicly accessible and proprietary resources. Examples of these supplementary datasets include DIARETDB1 [120] MESSIDOR [121], ARIA [122], REVIEW [123], GoDARTS (Genetics of Diabetes in Tayside Scotland: An Audit and Research), IOSTAR [124,125], and RCSLO.
Various techniques for classifying arteries and veins have been evaluated using VICAVR Vázquez, et al., (2013), INSPIREAVR [126], and WIDE [127]. Several databases, such as STARE, ARIA (Automated Retinal Image Analyzer), HRF, VAMPIRE, INSPIRE-AVR, and WIDE, incorporate images with pathological features like Diabetic Retinopathy (DR), Age-related Macular Degeneration (AMD), and glaucoma. This inclusion of images with lesions enables the testing of vessel detection methods’ robustness against potential confounding factors. In addition, databases like IOSTAR and RC-SLO tackle segmentation challenges related to Central Venous Refilling (CVR), fine vessels, and complex bifurcation or crossover points. Lastly, there are a handful of other datasets that are quite limited in size and therefore seldom employed in these evaluations.
In summary, a wide range of methods for retinal vessel segmentation have been analyzed using numerous databases, such as DRIVE, STARE, CHASEDB1, and HRF. Additional datasets have also been utilized, including public and proprietary ones like DIARETDB1, MESSIDOR, ARIA, REVIEW, GoDARTS, IOSTAR, and RCSLO. Artery and vein classification techniques have been evaluated through databases like VICAVR, INSPIREAVR, and WIDE. Moreover, these databases feature images with lesions, enabling robustness tests for vessel detection against confounding factors. Other databases, such as IOSTAR and RC-SLO, address specific segmentation issues related to CVR, thin vessels, and bifurcations or crossover points. However, a few datasets are quite small and rarely employed in these studies.
The scarcity of research into arterial and venular (A/V) division’s complexities, in comparison to the more widely examined domain of vessel segmentation, arises from multiple factors. A primary aspect is the importance of A/V labelling in statistical evaluations that seek to uncover correlations between retinal phenotype and clinical outcomes, requiring separate analysis of arterial and venular networks, which makes A/V labelling essential. However, not all entities involved in vessel classification participate in such studies, leading to limited access to A/V ground truth labels, as datasets containing this information are sparse and less prominent compared to vessel segmentation labels, which are easier to acquire and require only unlabelled vessel maps.
Deep diving into intricate medical images requires rigorous study of data sets displayed as blood vessels; however, parsing them becomes complicated when you include arterioles/venules (A/V). Though new players may overlook this section as it demands extra steps along with classifications; those relying on advanced deep learning solutions incorporating simultaneous segregation/ classification will find their tasks less cumbersome hence easier than ever before. With further innovation in this area & access being made easier to public datasets, we can expect to see more focus on A/V segmentation & classification in the future. To summarize, the discrepancy in the number of studies concentrating on A/V segmentation and vessel segmentation is due to the combined factors of restricted access to ground truth labels and the task’s added complexity. Nonetheless, with the advent of deep learning systems, the outlook for A/V segmentation research appears promising.
The precision of retinal vessel segmentation techniques has been a widely discussed topic in the research community, with some speculating if the limits of accuracy on datasets such as DRIVE and STARE have been attained. However, a closer examination of accuracy histograms reveals that there is still considerable scope for enhancement. Despite a clustering of accuracy around 95-96%, the range of accuracy stretches from 92%to98%, indicating a disparity in the accuracy achieved by various methods and, therefore, untapped potential for improvement.
The establishment of ground truth, the process of creating a reference standard for evaluating segmentation results, poses a significant obstacle in accurately determining the precision of retinal vessel segmentation methods. The medical image analysis community has not yet thoroughly tackled issues related to ground truth design, including annotator instructions, management of diverse annotations, the influence of ground truth design on specific objectives, the number of annotators, and evaluation criteria, among other concerns.
The DRIVE dataset exemplifies the difficulties connected to ground truth design. The creators of DRIVE split the 40 images into two subsets, with two manual annotators delineating the vasculature in one set and only one annotator in the other. This situation prompts questions about the impact of the number of annotators on accuracy and whether improving accuracy simply equates to aligning more closely with one or two annotators. This issue remains a subject of ongoing discussion within the medical image analysis community, as explored by Maier Hein, et al., [113] in their insightful article. In summary, although it may appear that the accuracy of retinal vessel segmentation has reached its zenith, there remains room for improvement, and ground truth design persists as a formidable challenge in accurately evaluating retinal vessel segmentation methods’ results. Further research and discussion are necessary to fully comprehend and address these challenges. Is it evident that Deep Learning (DL) excels over non-DL techniques in a quantitative sense? The solution is not as straightforward as one might assume. Without a doubt, DL has made significant progress in various domains, such as speech processing and generation, automated captioning, facial and object identification, retinal patient prioritization, and cardiovascular disease risk assessment [128]. At this juncture, it is reasonable to examine the extent of quantitative improvement in terms of performance benchmarks for the specific tasks involved, such as retinal blood vessel segmentation and classification.
The researchers endeavored to conduct a quantitative evaluation of the efficacy of both DL and non-DL approaches investigated for this objective. Nevertheless, this process is far from uncomplicated. To ensure a uniform evaluation, the focus was solely on methodologies assessed on DRIVE and STARE datasets, using accuracy as the lone reference standard. Consequently, a set of 17 DL and 33 non-DL research papers for DRIVE, as well as 13 DL and 25 non-DL papers for STARE, were analyzed during the time frame encompassed by this study.
This selection comprises 88 of the 158 papers incorporated into the review, signifying that uniformity is attained at the cost of comprehensiveness. In other words, to facilitate a fair comparison between DL and non-DL methods, the researchers had to narrow their focus to a specific set of papers, which limited the overall inclusiveness of their analysis. While deep learning has demonstrated undeniable advancements across various fields, determining its quantitative superiority over non-DL methods remains a complex task. The intricacies involved in conducting a fair and comprehensive quantitative analysis make it difficult to provide a simple answer to this question.
Histogram charts displaying the mean accuracy on DRIVE and STARE datasets were generated using a class-normalized distribution. Many studies provide distinct accuracies for healthy and pathological images; for the analysis, the accuracy of healthy images was employed. The histograms obtained for STARE are depicted in one figure, while those for DRIVE are presented in another. For the STARE dataset, the findings indicate that the majority of the examined DL techniques achieve considerably higher accuracy rates than most non-DL approaches, with DL methods attaining the highest overall accuracy (99% compared to 96%). Nevertheless, the situation with DRIVE is not as clear-cut. Unlike the STARE results, the DL and non-DL histograms for DRIVE are virtually indiscernible. The majority of both DL and non-DL methods reach peak accuracies in the range of 95-96% and 94-95%, respectively, which can be considered relatively close. In this case, non-DL techniques achieve the highest overall accuracy, albeit marginally higher than the best-performing DL method (99% versus 98%).
Thus, while DL methods demonstrate superior performance in the context of the STARE dataset, the distinction between DL and non-DL techniques is less evident when examining the DRIVE dataset. The results reveal a more nuanced picture, with both DL and non-DL methods achieving closely matched accuracy levels in certain instances. In light of the limitations of their analysis, which will be detailed in the subsequent paragraph, the researchers are inclined to surmise that Deep Learning (DL) has not yet displayed a definitive, substantial enhancement in accuracy for the specific challenge of retinal vessel detection using the DRIVE and STARE datasets. Owing to multiple constraints, this conclusion must be viewed as suggestive rather than definitive.
Firstly, as mentioned earlier, various research papers employ diverse datasets and criteria, complicating the process of conducting a fair comparison between algorithms. Secondly, and consequently, the sample size utilized in the analysis is smaller than what could have been possible. Thirdly, even when the same datasets and criteria are employed, testing protocols may differ (e.g., the number of folds, the division of data for training versus testing). DRIVE and STARE exhibit resolutions of 584x565 and 700x605, respectively, which are considerably lower than the resolutions offered by cutting- edge fundus cameras.
Nonetheless, the researchers consider this approach for two primary reasons. Firstly, DRIVE is the most commonly utilized benchmark dataset in the retinal image processing papers they examined, and newly published studies continue to incorporate it in their evaluations. Secondly, there is a need for publicly accessible, annotated image datasets with modern resolutions (approximately 3,000 pixels). While some of these datasets are beginning to emerge, such as through international challenges like MICCAI REFUGE or the High-Resolution Fundus (HRF) dataset, their prevalence in the literature is not yet on par with that of DRIVE.
In conclusion, within the boundaries of their analysis and given the numerous constraints, the researchers cautiously infer that deep learning has not yet demonstrated a significant improvement in accuracy for retinal vessel detection using the DRIVE and STARE datasets. However, it is crucial to acknowledge the limitations of the analysis, including the diverse datasets and criteria used across different papers, the smaller sample size, and varying testing protocols. These factors make it challenging to draw a definitive conclusion about the superiority of deep learning in retinal vessel detection. Furthermore, although the DRIVE and STARE datasets have lower resolutions than state-of-the-art fundus cameras, they remain the most commonly used benchmark datasets in retinal image processing research. To facilitate the further advancement of deep learning techniques in this field, there is a pressing need for publicly available, annotated image datasets with contemporary resolutions. Emerging datasets from international challenges, such as MICCAI REFUGE or HRF, may help bridge this gap and provide a more accurate representation of deep learning’s potential in retinal vessel detection.
Numerous algorithms, especially those employing high-accuracy deep learning techniques, often initiate the process by downsizing raw images to dimensions comparable to DRIVE, generally to decrease processing time. Consequently, the advantages of enhanced instrument resolution are substantially diminished or even completely nullified. This limitation, however, can be considered temporary: with the continuous advancements in accessible, affordable, and powerful computing platforms, this constraint will likely be overcome. Furthermore, the growth in computational power is not expected to be paralleled by an increase in image size, as fundus image resolutions are already approaching the maximum resolution permitted by the optical systems of fundus cameras. It is crucial to recognize that stating deep learning has no influence on retinal image analysis in general would be incorrect. The focus of this investigation has been on two highly specialized image processing tasks: vessel segmentation and classification. Deep learning has led to groundbreaking research on retinal biomarkers, demonstrating that the retina alone can predict personal traits and the presence of diseases with remarkable precision [128]. This field relates to modern artificial intelligence classification, which may or may not involve image processing techniques.
Additionally, the analysis was focused on fundus camera images; however, image analysis systems also exist for images captured using other devices, such as Scanning Laser Ophthalmoscopy (SLO), Optical Coherence Tomography (OCT), Optical Coherence Tomography Angiography (OCT-A), and autofluorescence. These instruments provide insights into various aspects and processes of the retina [129].
In summary, while certain algorithms downsize raw images to DRIVE-like dimensions, reducing the benefits of higher instrument resolution, this limitation is likely temporary due to advancements in computing platforms. Additionally, it is important to acknowledge that deep learning has a broader impact on retinal image analysis beyond the specific tasks of vessel segmentation and classification. Groundbreaking research in retinal biomarkers demonstrates the potential of deep learning to predict personal characteristics and disease presence. Finally, this analysis focused on fundus camera images, but other instruments such as SLO, OCT, OCT-A, and autofluorescence also provide valuable information about the retina and its various components, expanding the scope of retinal image analysis.
Conclusion
As medical diagnosis progresses with time so should the methods employed while examining key aspects of human anatomy correspondingly advance. One particular area that requires attention is the categorization as well as separation of retinal vessels into either an artery or vein category precisely defining the existence of crucial features that signify a specific illness such as cardiovascular disease, hypertension or even stroke among others when it comes to retinal and microvascular ailments. Distinguishing between the two categories of vessels, in which morphological differences are significant, is paramount. For instance, clinical evaluations and correlational studies might use measurements like Central Retinal Artery Equivalent (CRAE), Central Retinal Vein Equivalent (CRVE), or AVR to determine whether there are retinal indicators for various illnesses.
In recent years, the development of semi-automated and automated tools, including the Vascular Assessment and Measurement Platform for Retinal Images (VAMPIRE), SIVA, Interactive Vessel Analysis (IVAN), Quantitative Analysis of Retinal Vessel Topology (QUARTZ), and ARIA, has greatly improved the efficiency of retinal vessel examination. These advanced instruments calculate the morphological features of retinal vessels, enabling more accurate diagnosis and evaluation of retinal and microvascular disorders.
The current focus on machine learning and deep learning-driven methodologies signals a bright future for retinal vessel segmentation and categorization. As computational power advances and new high-resolution datasets emerge, it is expected that the reliability and precision of these systems will continue to improve, leading to enhanced patient outcomes. The accurate classification of retinal vessels into arteries and veins is crucial for diagnosing and evaluating retinal and microvascular conditions, making the ongoing refinement of these methods essential.
The emergence of highly accurate methodologies for discerning and categorizing retinal vessels via machine learning and deep learning relies upon the presence of copious annotated image data. Despite the abundance of imaging resources, such as the extensive UK Biobank housing over 50,000 retinal images, the endeavor to generate sufficient annotations presents a formidable challenge. A recent study by De Fauw, et al., [128] disclosed that the disparity between the number of images procured (1.5 million) and the volume of images that experts could annotate spanned 18 orders of magnitude.
Addressing this challenge is crucial for actualizing the broad automation of retinal vessel detection and classification. The conventional approach of manual annotation by clinicians, characterized by the laborious task of accentuating and encircling particular elements in images, is susceptible to errors and time-consuming, in addition to the risk of monotony. Alternatives that eliminate the necessity for annotations, such as appraising the performance of the entire system rather than a solitary module, may prove more efficacious. For instance, ascertaining the accuracy of a comprehensive DR screening instrument, as opposed to solely evaluating the performance of the microaneurysm detection module, delivers a more all-encompassing assessment of the system.
In conclusion, the establishment of dependable deep learning techniques for vessel detection and classification in retinal imaging necessitates the accessibility of an immense number of annotated images. The current constraints of publicly available data sets exemplify the difficulties intrinsic to annotating adequate quantities of images. Nevertheless, the pursuit of research targeting the reduction of annotation requirements harbors immense potential for the extensive automation of retinal vessel detection and classification. By examining the entire system, the demand for time-consuming and costly annotations can be diminished, yielding a more comprehensive evaluation of the system.
As the domain of retinal vessel segmentation and classification progresses, Non-Deep Learning (DL) methodologies may be instrumental in addressing a plethora of tasks. However, it is crucial to reassess the concept of what constitutes a “reasonably well-solved” problem and define it based on its real-world application rather than merely its performance on a restricted test set. The ultimate objective should be identifying the method that yields the most significant impact on healthcare outcomes, such as assisting in diagnosis, patient triage, or screening for Diabetic Retinopathy (DR), and contrasting it with other vessel classification and segmentation techniques in terms of advantages.
This change in viewpoint requires a move from solely assessing a method’s effectiveness based on specific validation metrics, such as the area under the receiver operating characteristic curve (AUC), to taking into account its actual impact on health outcomes in practice. For example, consider the possibility of using vessel-derived indicators to estimate the likelihood of developing systemic illnesses.
Assessing the effectiveness and worth of a technique on such an expansive scale is a considerably more extensive task than previously anticipated, demanding an in-depth comprehension of real- world healthcare applications and the various factors affecting a technique’s performance. As the global discourse on this subject continues to develop, it will be intriguing to observe how validation paradigms adjust to this novel perspective. Ultimately, the validation of a technique should concentrate on its impact on healthcare outcomes and its capability to enhance real-world healthcare applications [128,130-133].
Open Issues
Effective examination of the eyes complexities and understanding human vision has been made possible by implementing machine learning in ophthalmology. Retinal vessel segmentation was one task that had historically challenged medical practitioners but has now been successfully tackled through this advanced approach. Convolutional Neural Networks (CNN) provide solutions to intricate tasks such as image classification and object detection and form the basis of retinal vessel segmentation. Another technology contributing to this segmentations’ success is Conditional Random Fields (CRF) which models long distance pixel interactions necessary for accurately depicting retinal anatomy. The integration of these two powerful techniques created Deep Vessel, an innovative technology that merges CNN and CRF into a single deep network. Medical professionals now have access to a valuable instrument thanks to the system’s successful testing and validation using datasets like DRIVE, STARE, and CHASE. Automating retinal vessel segmentation has been a goal for the past two decades which led to various techniques being employed. Marin, et al., [134] utilized gray-plane vectors and moment invariant features for pixel classification via neural networks. Meanwhile, Nguyen, et al., [135] tackled retinal vessel segmentation through multi-scale line detection and assessed vessel fragmentation. In summary, the incorporation of machine learning into ophthalmology has revolutionized the challenging process of segmenting retinal vessels.
The innovative Deep Vessel system, combining the strengths of CNN and CRF, exemplifies how technology can revolutionize medical processes, enhancing efficiency and accuracy. As technology continues to progress, we can anticipate the emergence of more sophisticated solutions, driving the field of ophthalmology forward.
The progress of electronic vision has attained new milestones in recent years, particularly with the incorporation of Deep Learning (DL) techniques. Trailblazing researchers like Nguyen and Or lando have made significant contributions to the field, employing a multi-scale line detection approach and a fully connected Conditional Random Field (CRF) respectively, for achieving retinal vessel segmentation. However, these methods still fall short in providing distinct representations, rendering them susceptible to interference from pathological regions, thus affecting the quality of their segmentation performance.
Conversely, the potency of DL in delivering clear and distinguishable representations has been well established, with Convolutional Neural Networks (CNNs) being especially noteworthy for their proficiency in image classification and semantic segmentation. By outperforming traditional techniques, DL has paved the way for solving numerous issues in the realm of electronic vision.
A standout illustration of DL’s representation capabilities is the work of Xie, et al., [136], who utilized deep-edge total edge detection (HED) systems to clarify ambiguous boundaries in object detection. The unique representations generated by HED have enabled it to produce remarkably accurate results across various scenarios [137-140].
In summary, conventional methods for retinal vessel segmentation, despite their merits, are limited by their inability to provide distinct representations, making them vulnerable to interference from pathological regions. On the other hand, DL has proven to deliver highly distinct representations, making it a more fitting solution to address the challenges presented by electronic vision, including retinal vessel segmentation [141-143].
Acknowledgement
None.
Open Issues
None.
References
- Fu H, Xu Y, Lin S, Kee Wong DW, Liu J, et al. (2016) Deepvessel: Retinal vessel segmentation via deep learning and conditional random field. In Medical Image Computing and Computer-Assisted Intervention-MICCAI 2016: 19th International Conference, Athens, Greece, October: 17-21. Proceedings, Part II 19 132-139. Springer International Publishing.
- Ding L, Bawany MH, Kuriyan AE, Ramchandran RS, Wykoff CC, et al. (2020) A novel deep learning pipeline for retinal vessel detection in fluorescein angiography. IEEE Transactions on Image Processing 29: 6561-6573.
- Yan Z, Yang X, Cheng KT (2019) A three-stage deep learning model for accurate retinal vessel segmentation. IEEE J Biomed Health Inform 23(4): 1427-1436.
- Wang L, Liu H, Lu Y, Chen H, Zhang J, et al. (2019) A coarse-to-fine deep learning framework for optic disc segmentation in fundus images. Biomed Signal process Control 51: 82-89.
- Xiuqin P, Zhang Q, Zhang H, Li S (2019) A fundus retinal vessels segmentation scheme based on the improved deep learning U-Net model. IEEE Access 7: 122634-122643.
- Wu C, Zou Y, Zhan J (2019) DA-U-Net: densely connected convolutional networks and decoder with attention gate for retinal vessel segmentation. In IOP Conference Series: Materials Science and Engineering 533: 012053.
- Zhang B, Zhang L, Zhang L, Karray F (2010) Retinal vessel extraction by matched filter with first-order derivative of Gaussian. Comput Biol Med 40(4): 438-445.
- Vlachos M, Dermatas E (2010) Multi-scale retinal vessel segmentation using line tracking. Compute Med Imaging Graph 34(3): 213-227.
- Espona L, Carreira MJ, Penedo MG, Ortega M (2008) Retinal vessel tree segmentation using a deformable contour model. In 2008 19th International Conference on Pattern Recognition pp.1-4.
- Soomro TA, Afifi AJ, Zheng L, Soomro S, Gao J, et al. (2019) Deep learning models for retinal blood vessels segmentation: a review. IEEE Access 7: 71696-71717.
- Rodrigues EO, Conci A, Liatsis P (2020). ELEMENT: Multi-modal retinal vessel segmentation based on a coupled region growing and machine learning approach. IEEE J Biomed Health Inform 24(12): 3507-3519.
- Maninis KK, Pont Tuset J, Arbeláez P, Van Gool L (2016) Deep retinal image understanding. Proceedings pp.140-148.
- Ghosh TK, Saha S, Rahaman GA, Sayed MA, Kanagasingam Y (2019) Retinal blood vessel segmentation: A semi-supervised approach. In Pattern Recognition and Image Analysis: 9th Iberian Conference, Proceedings, Part II 9: 98-107.
- Pissas T, Bloch E, Cardoso MJ, Flores B, Georgiadis O, et al. (2020) Deep iterative vessel segmentation in OCT angiography. Biomed Opt Express 11(5): 2490-2510.
- Ryu H, Moon H, Browatzki B, Wallraven C (2018) Retinal vessel detection using deep learning: a novel Directnet architecture. Korean J Vis Sci 20(2): 151-159.
- Prentašić P, Heisler M, Mammo Z, Lee S, Merkur A, et al. (2016) Segmentation of the foveal microvasculature using deep learning networks. J biomed opt 21(7): 075008-075008.
- Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, et al. (2016) Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 316(22): 2402-2410.
- Mookiah MRK, Hogg S, MacGillivray TJ, Prathiba V, Pradeepa R, et al. (2021) A review of machine learning methods for retinal blood vessel segmentation and artery/vein classification. Med Image Anal 68: 101905.
- Fraz MM, Remagnino P, Hoppe A, Uyyanonvara B, Rudnicka AR, et al. (2012) Blood vessel segmentation methodologies in retinal images a survey. Computer methods and programs in biomedicine 108(1): 407-433.
- L Srinidhi C, Aparna P, Rajan J (2017) Recent advancements in retinal vessel segmentation. J med sys 41(4): 1-22.
- Wang S, Yin Y, Cao G, Wei B, Zheng Y, et al. (2015) Hierarchical retinal blood vessel segmentation based on feature and ensemble learning. Neurocomputing 149: 708-717.
- Tan J, Acharya UR, Bhandary SV, Chua KC, Sivaprasad S (2017) Segmentation of optic disc, fovea and retinal vasculature using a single convolutional neural network. Journal of Computational Science 20: 70-79.
- Luo Y, Yang L, Wang L, Cheng H (2017) Efficient CNN-CRF network for retinal image segmentation. Cognitive Systems and Signal Processing pp.157-165.
- Liskowski P, Krawiec K (2016) Segmenting retinal blood vessels with deep neural networks. IEEE trans med imaging 35(11): 2369-2380.
- Yan Z, Yang X, Cheng KT (2018) Joint segment-level and pixel-wise losses for deep learning based retinal vessel segmentation. IEEE Trans Biomed Eng 65(9): 1912-1923.
- Oliveira A, Pereira S, Silva CA (2018) Retinal vessel segmentation based on fully convolutional neural networks. Expert Systems with Applications 112: 229-242.
- Franklin SW, Rajan SE (2014) Computerized screening of diabetic retinopathy employing blood vessel segmentation in retinal images. biocybernetics and biomedical engineering 34(2): 117-124.
- Rahebi, J, Hardalaç F (2014) Retinal blood vessel segmentation with neural network by using gray-level co-occurrence matrix-based features. J med syst 38(8): 85.
- Vega R, Sanchez Ante G, Falcon Morales LE, Sossa H, et al. (2015) Retinal vessel extraction using lattice neural networks with dendritic processing. Comput Biol Med 58: 20-30.
- Fraz MM, Rudnicka AR, Owen CG, Barman SA (2014) Delineation of blood vessels in pediatric retinal images using decision trees-based ensemble classification. International journal of computer assisted radiology and surgery 9: 795-811.
- Fathi A, Naghsh Nilchi AR (2014) General rotation-invariant local binary patterns operator with application to blood vessel detection in retinal images. Pattern Analysis and Applications 17: 002069-002081.
- Cao S, Bharath AA, Parker KH, Ng J (2012) Patch-based automatic retinal vessel segmentation in global and local structural context. Annual International Conference of the IEEE Engineering in Medicine and Biology Society 2012: 4942-4945.
- Shah SAA, Tang TB, Faye I, Laude A (2017) Blood vessel segmentation in color fundus images based on regional and Hessian features. Graefe's Arch Clin Exp Ophthalmol 255(8): 1525-1533.
- Zhang L, Fisher M, Wang W (2015) Retinal vessel segmentation using multi-scale textons derived from keypoints. Comput Med Imaging Graph 45: 47-56.
- Annunziata R, Trucco E (2016) Accelerating convolutional sparse coding for curvilinear structures segmentation by refining SCIRD-TS filter banks. IEEE transactions on medical imaging 35(11): 2381-2392.
- Memari N, Ramli AR, Bin Saripan MI, Mashohor S, Moghbel M (2017) Supervised retinal vessel segmentation from color fundus images based on matched filtering and AdaBoost classifier. PloS one 12(12): e0188939.
- Geetha Ramani R, Balasubramanian L (2016) Retinal blood vessel segmentation employing image processing and data mining techniques for computerized retinal image analysis. Biocybernetics and Biomedical Engineering 36(1): 102-118.
- Fathi A, Naghsh Nilchi AR (2013) Integrating adaptive neuro-fuzzy inference system and local binary pattern operator for robust retinal blood vessels segmentation. Neural Computing and Applications 22: 163-174.
- Sigurðsson EM, Valero S, Benediktsson JA, Chanussot J, Talbot H, et al. (2014) Automatic retinal vessel extraction based on directional mathematical morphology and fuzzy classification. Pattern Recognition Letters 47: 164-171.
- Zhang B, Karray F, Li Q, Zhang L (2012) Sparse representation classifier for microaneurysm detection and retinal blood vessel extraction. Information Sciences 200: 78-90.
- Zhu C, Zou B, Zhao R, Cui J, Duan X, et al. (2017) Retinal vessel segmentation in colour fundus images using extreme learning machine. Comput Med Imaging Graph 55: 68-77.
- Zhu C, Zou B, Xiang Y, Cui J, Wu H (2016) An ensemble retinal vessel segmentation based on supervised learning in fundus images. Chinese Journal of Electronics 25(3): 503-511.
- Orlando JI, Prokofyeva E, Blaschko MB (2016) A discriminatively trained fully connected conditional random field model for blood vessel segmentation in fundus images. IEEE trans Biomed Eng 64(1): 16-27.
- Orlando JI, Blaschko M (2014) Learning fully-connected CRFs for blood vessel segmentation in retinal images. Med Image Comput Comput Assist Interv 17(1): 634-641.
- Ganjee R, Azmi R, Gholizadeh B (2014) An improved retinal vessel segmentation method based on high level features for pathological images. Journal of medical systems 38(9): 108.
- Waheed A, Akram MU, Khalid S, Waheed Z, Khan MA, et al. (2015) Hybrid features and mediods classification based robust segmentation of blood vessels. J Med Syst 39(10): 128.
- Tang Z, Zhang J, Gui W (2017) Selective search and intensity context-based retina vessel image segmentation. J Med Syst 41(3): 47.
- Panda R, Puhan NB, Panda G (2016) New binary Hausdorff symmetry measure based seeded region growing for retinal vessel segmentation. Biocybernetics and Biomedical Engineering 36(1): 119-129.
- Ding Y, Ward WO, Duan J, Auer DP, Gowland P, et al. (2015) Retinal vasculature classification using novel multifractal features. Physics in Medicine & Biology 60(21): 8365-8379.
- Strisciuglio N, Azzopardi G, Vento M, Petkov N (2016) Supervised vessel delineation in retinal fundus images with the automatic selection of B-COSFIRE filters. Machine Vision and Applications 27: 1137-1149.
- Srinidhi CL, Aparna P, Rajan J (2018) A visual attention guided unsupervised feature learning for robust vessel delineation in retinal images. Biomedical Signal Processing and Control 44: 110-126.
- Cheng E, Du L, Wu Y, Zhu YJ, Megalooikonomou V, et al. (2014) Discriminative vessel segmentation in retinal images by fusing context-aware hybrid features. Machine vision and applications 25: 1779-1792.
- Roychowdhury S, Koozekanani DD, Parhi KK (2015) Blood vessel segmentation of fundus images by major vessel extraction and subimage classification. IEEE J B Health Inform 19(3): 1118-1128.
- Emary E, Zawbaa HM, Hassanien AE, Parv B (2017) Multi-objective retinal vessel localization using flower pollination search algorithm with pattern search. Advances in data analysis and classification 11: 611-627.
- Hassanien AE, Emary E, Zawbaa HM (2015) Retinal blood vessel localization approach based on bee colony swarm optimization, fuzzy c-means and pattern search. Journal of Visual Communication and Image Representation 31: 186-196.
- Hassan G, Hassanien AE (2018) Retinal fundus vasculature multilevel segmentation using whale optimization algorithm. Signal, Image and Video Processing 12(2): 263-270.
- Neto LC, Ramalho GL, Neto JFR, Veras RM, Medeiros FN (2017) An unsupervised coarse-to-fine algorithm for blood vessel segmentation in fundus images. Expert Systems with Applications 78: 182-192.
- Chaudhuri S, Chatterjee S, Katz N, Nelson M, Goldbaum M, et al. (1989) Detection of blood vessels in retinal images using two-dimensional matched filters. IEEE Transactions on medical imaging 8(3): 263-269.
- Kovács G, Hajdu A (2016) A self-calibrating approach for the segmentation of retinal vessels by template matching and contour reconstruction. Med image anal 29: 24-46.
- Singh NP, Srivastava R (2016) Retinal blood vessels segmentation by using gumbel probability distribution function based matched filter. Comput Methods Programs Biomed 129: 40- 50.
- Liu Q, Zou B, Chen J, Chen Z, Zhu C, et al. (2016) Retinal vessel segmentation from simple to difficult. Proceedings of the Ophthalmic Medical Image Analysis Third International Workshop.
- Kar SS, Maity SP (2016) Retinal blood vessel extraction using tunable bandpass filter and fuzzy conditional entropy. Comput methods and programs in biomed 133: 111-132.
- Kar SS, Maity SP (2016) Retinal blood vessel extraction and optic disc removal using curvelet transform and morphological operation. Machine Intelligence and Signal Processing: 153-161.
- Kar SS, Maity SP (2016) Blood vessel extraction and optic disc removal using curvelet transform and kernel fuzzy c-means. Comput biol med 70: 174-189.
- Tan JH, Acharya UR, Chua KC, Cheng C, Laude A (2016) Automated extraction of retinal vasculature. Med phys 43(5): 2311- 2322.
- Soomro TA, Khan MAU, Gao J, Khan TM, Paul M (2017) Contrast normalization steps for increased sensitivity of a retinal image segmentation method. Signal, Image and Video Processing 11: 1509-1517.
- Rezaee K, Haddadnia J, Tashk A (2017) Optimized clinical segmentation of retinal blood vessels by using combination of adaptive filtering, fuzzy entropy and skeletonization. Applied Soft Computing 52: 937-951.
- Subudhi A, Pattnaik S, Sabut S (2016) Blood vessel extraction of diabetic retinopathy using optimized enhanced images and matched filter. Journal of Medical Imaging 3(4): 044003.
- Farokhian F, Yang C, Demirel H, Wu S, Beheshti I, et al. (2017) Automatic parameters selection of gabor filters with the imperialism competitive algorithm with application to retinal vessel segmentation. Biocybernetics and Biomedical Engineering 37(1): 246-254.
- Koukounis D, Ttofis C, Papadopoulos A, Theocharides T (2014) A high-performance hardware architecture for portable, low-power retinal vessel segmentation. INTEGRATION, the VLSI journal 47(3): 377-386.
- Krause M, Alles RM, Burgeth B, Weickert J (2016) Fast retinal vessel analysis. Journal of Real-Time Image Processing 11(2): 413-422.
- Haralick RM, Sternberg SR, Zhuang X (1987) Image analysis using mathematical morphology. IEEE transactions on pattern analysis and machine intelligence (4): 532-550.
- Badsha S, Reza AW, Tan KG, Dimyati K (2013) A new blood vessel extraction technique using edge enhancement and object classification. Journal of digital imaging 26(6): 1107-1115.
- Fraz MM, Basit A, Barman S (2013) Application of morphological bit planes in retinal blood vessel extraction. Journal of digital imaging 26 (2): 274-286.
- Saleh MD, Eswaran C (2012) An efficient algorithm for retinal blood vessel segmentation using h-maxima transform and multilevel thresholding. Comput Methods Biomech Biomed Engin 15(5): 517-525.
- Imani E, Javidi M, Pourreza HR (2015) Improvement of retinal blood vessel detection using morphological component analysis. Computer methods and programs in biomedicine 118(3): 263-279.
- Jiang Z, Yepez J, An S, Ko S (2017) Fast, accurate and robust retinal vessel segmentation system. Biocybernetics and Biomedical Engineering 37(3): 412-421.
- Hassan M, Amin M, Murtza I, Khan A, Chaudhry A, et al. (2017) Robust hidden markov model based intelligent blood vessel detection of fundus images. Computer Methods and Programs in Biomedicine 151: 193-201.
- Dash J, Bhoi N (2017) A thresholding-based technique to extract retinal blood vessels from fundus images. Future Computing and Informatics Journal 2 (2): 103-109.
- Nayebifar B, Moghaddam HA (2013) A novel method for retinal vessel tracking using particle filters. Comput biol med 43(5): 541-548.
- Lin KS, Tsai CL, Tsai CH, Sofka M, Chen SJ, et al. (2012) Retinal vascular tree reconstruction with anatomical realism. IEEE Transactions on Biomedical Engineering 59(12): 3337-3347.
- Yang X, Liu C, Le Minh H, Wang Z, Chien A, et al. (2017) An automated method for accurate vessel segmentation. Physics in Medicine & Biology 62 (9): 3757.
- Wang L, Kallem V, Bansal M, Eledath J, Sawhney H, et al. (2013) Interactive retinal vessel extraction by integrating vessel tracing and graph search. Med Image Comput Comput Assist Interv 16(Pt 2): 567- 574.
- Sofka M, Stewart CV (2006) Retinal vessel centerline extraction using multiscale matched filters, confidence and edge measures. IEEE Trans Med Imaging 25(12): 1531-1546.
- Bekkers E, Duits R, Berendschot T, terHaarRomeny B (2014) A multi-orientation analysis approach to retinal vessel tracking. Journal of Mathematical Imaging and Vision 49(3): 583-610.
- Nergiz M, Akin M (2017) Retinal vessel segmentation via structure tensor coloring and anisotropy enhancement. Symmetry 9(11): 276.
- Khan KB, Khaliq AA, Jalil A, Shahid M (2018) A robust technique based on vlm and frangi filter for retinal vessel extraction and denoising. PloS one 13(2): e0192203.
- Welikala R, Foster P, Whincup P, Rudnicka A, Owen C, et al. (2017) Automated arteriole and venule classification using deep learning for retinal images from the UK biobank cohort. Comput Biol Med 90: 23-32.
- Girard F, Cheriet F (2017) Artery/vein classification in fundus images using cnn and likelihood score propagation. In: 2017 IEEE Global Conference on Signal and Information Processing (GlobalSIP). IEEE pp.720-724.
- Girard F, Kavalec C, Cheriet F (2019) Joint segmentation and classification of retinal arteries/veins from fundus images. Artificial intelligence in medicine 94: 96-109.
- Hemelings R, Elen B, Stalmans I, Van Keer K, De Boever P, et al. (2019) Artery vein segmentation in fundus images using a fully convolutional network. Computerized Medical Imaging and Graphics 76: 101636.
- Ma W, Yu S, Ma K, Wang J, Ding X, et al. (2019) Multi-task neural networks with spatial activation for retinal vessel segmentation and artery/vein classification. International Conference on Medical Image Computing and Computer-Assisted Intervention pp.769-778.
- Yang J, Dong X, Hu Y, Peng Q, Tao G, et al. (2020) Fully automatic arteriovenous segmentation in retinal images via topology-aware generative adversarial networks. Interdiscip Sci 12(3): 323-334.
- Dashtbozorg B, Mendona AM, Campilho A (2013) Automatic estimation of the arteriolar-to-venular ratio in retinal images using a graph-based approach for artery/vein classification. In: International Conference Image Analysis and Recognition. Springer pp.530-538.
- Dashtbozorg B, Mendon a AM, Campilho AJ (2014) An automatic graph-based approach for artery/vein classification in retinal images. IEEE Trans. Image Processing 23(3): 1073-1083.
- Huang F, Dashtbozorg B, terHaarRomeny BM (2018) Artery/vein classification using re ection features in retina fundus images. Machine Vision and Applications 29(1): 23-34.
- Pellegrini E, Robertson G, MacGillivray T, van Hemert J, Houston G, et al. (2018) A graph cut approach to artery/vein classification in ultra-widefield scanning laser ophthalmoscopy. IEEE trans med imaging 37(2): 516-526.
- Akbar S, Akram MU, Sharif M, Tariq A, ullah Yasin U, et al. (2018) Arteriovenous ratio and papilledema-based hybrid decision support system for detection and grading of hypertensive retinopathy. Computer methods and programs in biomedicine 154: 123- 141.
- Hu Q, Abrmo MD, Garvin MK (2015) Automated construction of arterial and venous trees in retinal images. Journal of Medical Imaging 2(4): 044001.
- Vapnik V, Guyon I, Hastie T (1995) Support vector machines. Mach. Learn 20(3): 273- 297.
- Estrada R, Allingham MJ, Mettu PS, Cousins SW, Tomasi C, et al. (2015) Retinal artery-vein classification via topology estimation. IEEE transactions on medical imaging 34(12): 2518-2534.
- Xu X, Ding W, Abràmoff MD, Cao R (2017) An improved arteriovenous classification method for the early diagnostics of various diseases in retinal image. Comput Methods Programs Biomed 141: 3-9.
- Joshi VS, Reinhardt JM, Garvin MK, Abramo MD (2014) Automated method for identification and artery-venous classification of vessel trees in retinal vessel networks. PloS one 9(2): e88061.
- Yin X, Irshad S, Zhang Y (2020) Classifiers fusion for improved vessel recognition with application in quantification of generalized arteriolar narrowing. Journal of Innovative Optical Health Sciences 13(01): 1950021.
- Fu D, Liu Y, Ma H (2017) An artery/vein classification method based on color and vascular structure information. In: Lecture Notes in Computer Science. Springer International Publishing pp.487-495.
- Relan D, MacGillivray T, Ballerini L, Trucco E (2013) Retinal vessel classification: sorting arteries and veins. Annu Int Conf IEEE Eng Med Biol Soc 2013: 7396-7399.
- Relan D, Ballerini L, Trucco E, MacGillivray T (2019) Using orthogonal locality preserving projections to find dominant features for classifying retinal blood vessels. Multimedia Tools and Applications 78(10): 12783-12803.
- Yan Y, Wen D, Dewan MAA, Huang WB (2017) Classification of artery and vein in retinal fundus images based on the context-dependent features. In: International Conference on Digital Human Modeling and Applications in Health, Safety, Ergonomics and Risk Management pp.198-213.
- Lau QP, Lee ML, Hsu W, Wong TY (2013) Simultaneously identifying all true vessels from segmented retinal images. IEEE trans biomed eng 60(7): 1851-1858.
- Eppenhof K, Bekkers E, Berendschot TT, Pluim JP, terHaarRomeny BM, et al. (2015) Retinal artery/vein classification via graph cut optimization. In: Proceedings of the Ophthalmic Medical Image Analysis Second International Workshop. University of Iowa.
- Srinidhi CL, Aparna P, Rajan J (2019) Automated method for retinal artery/vein separation via graph search metaheuristic approach. IEEE Trans Image Process 28(6): 2705-2718.
- Mahum R, Rehman SU, Okon OD, Alabrah A, Meraj T, et al. (2021) A novel hybrid approach based on deep CNN to detect glaucoma using fundus imaging. Electronics 11(1): 26.
- Maier Hein L, Eisenmann M, Reinke A, Onogur S, Stankovic, et al. (2018) Why rankings of biomedical image analysis competitions should be interpreted with care. Nature communications 9(1): 5217.
- Joskowicz L, Cohen D, Caplan N, Sosna J (2019) Inter-observer variability of manual contour delineation of structures in ct. Eur radiol 29(3): 1391-1399.
- Trucco E, Ruggeri A, Karnowski T, Giancardo L, Chaum E, et al. (2013) Validating retinal fundus image analysis algorithms: issues and a proposal. Investigative ophthalmology & visual science 54(5): 3546-3559.
- Silberzahn R, Uhlmann EL (2015) Crowdsourced research: Many hands make tight work. Nature 526 (7572): 189-191.
- Hoover A, Kouznetsova V, Goldbaum M (2000) Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Transactions on Medical imaging 19(3): 203-210.
- Owen CG, Rudnicka AR, Mullen R, Barman SA, Monekosso D, et al. (2009) Measuring retinal vessel tortuosity in 10-year-old children: validation of the computer-assisted image analysis of the retina (caiar) program. Invest ophthalmol vis sci 50(5): 2004-2010.
- Budai A, Bock R, Maier A, Hornegger J, Michelson G, et al. (2013) Robust vessel segmentation in fundus images. International journal of biomedical imaging 2013: 154860.
- Kälviäinen RVJPH, Uusitalo H, T Kauppi, V Kalesnykiene, K Kamarainen, et al. (2007) DIARETDB1 diabetic retinopathy database and evaluation protocol. In Medical image understanding and analysis: 61.
- Decencière E, Zhang X, Cazuguel G, Lay B, Cochener B, et al. (2014) Feedback on a publicly distributed database: the messidor database. Image Analysis & Stereology 33(3): 231-234.
- Zheng Y, Hijazi MHA, Coenen F (2012) Automated disease-no disease grading of age-related macular degeneration by an image mining approach. Invest Ophthalmol Vis Sci 53(13): 8310-8318.
- Al Diri B, Hunter A, Steel D, Habib M, Hudaib T, et al. (2008) A reference data set for retinal vessel pro les. In: 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society 2008: 2262-2265.
- Zhang J, Dashtbozorg B, Bekkers E, Pluim JP, Duits R, et al. (2016) Robust retinal vessel segmentation via locally adaptive derivative frames in orientation scores. IEEE Trans Med Imaging 35(12), 2631-2644.
- Perez Rovira A, Zutis K, Hubschman JP, Trucco E (2011) Improving vessel segmentation in ultrawide eld-of-view retinal fluorescein angiograms. Annu Int Conf IEEE Eng Med Biol Soc 2011: 2614-2617.
- Niemeijer M, Xu X, Dumitrescu AV, Gupta P, Van Ginneken, et al. (2011) Automated measurement of the arteriolar-to-venular width ratio in digital color fundus photographs. IEEE Trans med imaging 30(11): 1941-1950.
- Estrada R, Tomasi C, Schmidler SC, Farsiu S (2015) Tree topology estimation. IEEE transactions on pattern analysis and machine intelligence 37(8): 1688-1701.
- De Fauw J, Ledsam JR, Romera Paredes B, Nikolov S, Tomasev N, et al. (2018) Clinically applicable deep learning for diagnosis and referral in retinal disease. Nature medicine 24(9): 1342.
- Mookiah MRK, Acharya UR, Fujita H, Tan JH, Chua CK, et al. (2015) Application of different imaging modalities for diagnosis of diabetic macular edema: a review. Computers in biology and medicine 66: 295-315.
- Valindria VV, Lavdas I, Bai W, Kamnitsas K, Aboagye EO, et al. (2017) Reverse classification accuracy: predicting segmentation performance in the absence of ground truth. IEEE Trans Med Imaging 36(8): 1597-1606.
- Joyce T, Chartsias A, Tsaftaris SA (2018) Deep multi-class segmentation without ground-truth labels.
- Huo Y, Xu Z, Moon H, Bao S, Assad A, et al. (2018) Synseg-net: Synthetic segmentation without target modality ground truth. IEEE transactions on medical imaging.
- Moccia S, De Momi E, El Hadji S, Mattos LS (2018) Blood vessel segmentation algorithms-review of methods, datasets and evaluation metrics. Comput methods programs biomed 158: 71-91.
- Marín D, Aquino A, Gegúndez Arias ME, Bravo JM (2010) A new supervised method for blood vessel segmentation in retinal images by using gray-level and moment invariants-based features. IEEE Transactions on medical imaging 30(1): 146-158.
- Nguyen UT, Bhuiyan A, Park LA, Ramamohanarao K (2013) An effective retinal blood vessel segmentation method using multi-scale line detection. Pattern recognition 46(3): 703-715.
- Xie S, Tu Z (2015) Holistically nested edge detection. In Proceedings of the IEEE international conference on computer vision pp.1395-1403.
- Citeseer Kalaie S, Gooya A (2017) Vascular tree tracking and bifurcation points detection in retinal images using a hierarchical probabilistic model. Computer methods and programs in biomedicine 151: 139-149.
- Dai L, Wu L, Li H, Cai C, Wu Q, et al. (2021) A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nature communications 12(1): 3242.
- Gonzalez RC, Woods RE (2007) Image processing. Digital image processing: 2(1): 1979.
- Lo SC, Lou SL, Lin JS, Freedman MT, Chien MV, et al. (1995) Artificial convolution neural network techniques and applications for lung nodule detection. IEEE trans med imaging 14(4): 711-718.
- Mahapatra D (2016) Retinal image quality classification using neurobiological models of the human visual system. Ophthalmic Medical Image Analysis International Workshop 3(2016): 97-104.
- (2007) Medical Image Understanding and Analysis. Citeseer pp.61.
- Mo J, Zhang L (2017) Multi-level deep supervised networks for retinal vessel segmentation. Int j comput assist surg 12(12): 2181-2193.



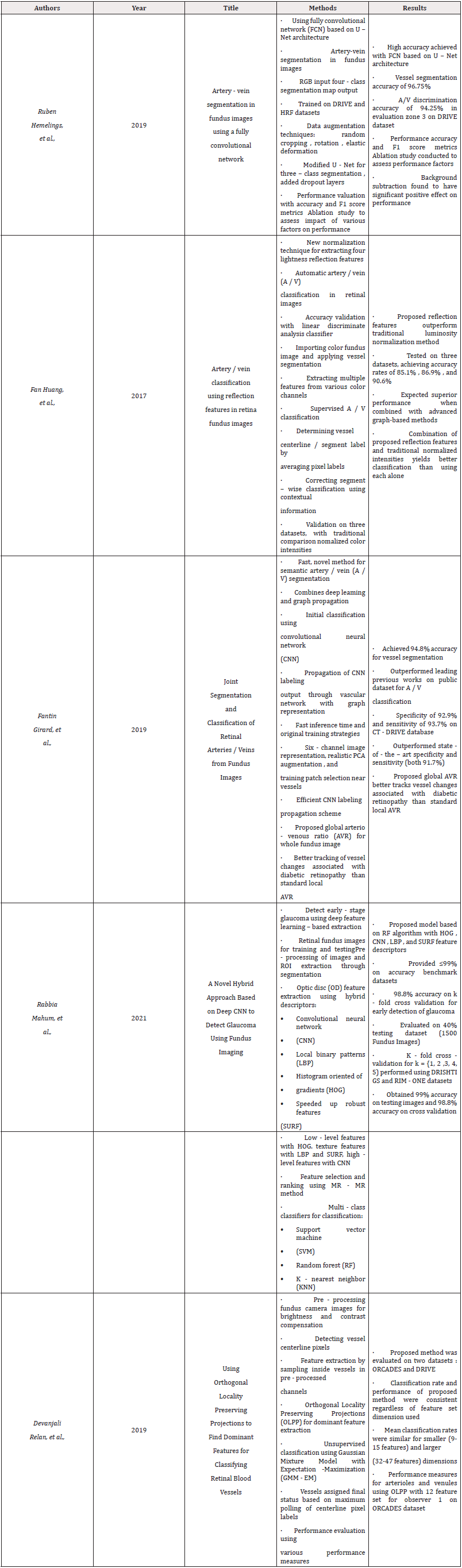


 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.