Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Using Bioinformatics to Study Homology Between Herpes Simplex Virus Type 1 Glycoprotein C Gene and Herpes Simplex Virus Type 2 Glycoprotein F Gene
*Corresponding author: Shereen F Elkholy, Department of Biology, College of Science and Humanities, Prince Sattam bin Abdulaziz University, Alkharj, Kingdom of Saudi Arabia
Received: February 19, 2019 Published: March 29, 2019
DOI: 10.34297/AJBSR.2019.02.000575
Abstract
Bioinformatics is an interdisciplinary field mainly involving molecular genetics. computer science, mathematics, and statistics. Data intensive and large-scale biological problems are addressed from a computational point of view. The most common problems are modeling biological processes at the molecular level and making inferences from collected data. In this study we used bioinformatics as a tool to study homology between Herpes simplex virus type 1 glycoprotein C gene and Herpes simplex virus type 2glycoprotein F gene. In the beginning we used the Gene data Bank to get the DNA sequence. Then we used a program Blast and got a percentage alignment 81%. We used protein data bank (PDB) to estimate the structure of the expressed proteins that were tertiary (three dimensional) structure that harbor both α helices and β pleated sheets. Finally, we used “PHYRE2 “protein folding recognition set to visualize, analyze and manipulate the resulted 3D protein model.
Introduction
Bioinformatics is conceptualizing Biology in terms of molecules and applying “informatics techniques” (derived from disciplines such as applied math’s, computer science and statistics) to understand and organize the information associated with these molecules, on a large scale. In short, bioinformatics is a management information system for molecular biology and has many practical applications. Bioinformatics is an application of techniques from computer science to problems from Biology. It is link between Computer Science and Biology. And has three aims: the first one: organizes data in a way that allows researchers to access existing information and to submit new entries as they are produced, e.g. the Protein Data bank for 3D macromolecular structures. The second aim: is to develop tools and resources that aid in the analysis of data. For example, having sequenced a protein, it is of interest to compare it with previously characterized sequences. The third aim: is to use these tools to analyze the data and interpret the results in a biologically meaningful manner.
Bioinformatics is a very broad field and it encompasses issues like mapping, sequencing, sequence comparison, gene identification, protein modeling, network databases, visualization and ethics. It is an interdisciplinary subject that on one hand requires biological information-infrastructure building and on the other requires computation based biological research. All this depends on the large stores of experimental and derived data.
The foundation of Bioinformatics is based on the computational techniques, algorithms, artificial intelligence, database management, software engineering etc. All this leads to the development of community data resources and from this starts its applications development of the bioinformatics for analysis of the genetic data.
In this study, we used Bioinformatics to detect the homology and the percentage of alignment between Herpes simplex virus type 1 glycoprotein C gene and Herpes simplex virus type 2glycoprotein F gene.
Herpes simplex virus
Is an enveloped double stranded DNA virus which encodes information for at least 10 glycoproteins. Three of the glycoproteins have functions which may modulate the immune response. In HSV- 1, glycoprotein E (gE) and gI function as a complex to bind the Fc portion of immunoglobulin G [3,4,6,7,15,6,22]. gC binds the C3b and iC3b fragments of the third component of complement [7-9,14,20,21,26-28]. Although gC is not strictly required for infection in cell culture [5,12,43,17,29], it plays an important role in virus attachment [11,25].
More recently, in-frame deletion and linker insertion mutants of gC-2 were used to identify regions important for C3b binding [26,27]. These studies showed that amino-terminal residues 26 through 73 of gC-2 are not involved in C3b receptor activity. In addition, three distinct regions (I, II, and III) in gC-2 are important for C3b binding. Region III has features like those of the short consensus repeat (SCR) [13,18,23,24,28] of the human C3-C4 receptor CR1 [26].
Several studies employed a transient transfection system using the gC-2 gene cloned into the genomes of herpes simplex virus type 1 (HSV-1) and HSV-2 encode at least four different glycoproteins, gA/B, gC, gD, and gE, which are found on the surface of the infected cell and the virion [39]. Three of the four glycoproteins, gA/B, gD, and gE, have been found to be structurally similar in the two virus types, based on immunological and biochemical criteria [33-37]. For example, a recent analysis of the DNA sequences of the gD genes from HSV-1 and HSV-2 revealed that the gD proteins had an overall sequence homology of 85% (L. Lasky and D. Dowbenko, DNA, in press). Thus, it may be concluded that the primary sequences of these three glycoproteins have been relatively well conserved, since the two virus types diverged from each other. HSV-1 gC initially appeared to have no obvious homolog in HSV-2. HSV-1 gC was thought to be type specific since antibodies against this glycoprotein were found to react almost exclusively with HSV-1 gC [38]. In addition, no detectable immunological reactions could be demonstrated between HSV-1 gC and antisera made against HSV-2 [32]. A protein having the same electrophoretic mobility as HSV- 1 gC has been demonstrated in HSV-2; however, it did not map collinearly with HSV-1 gC [17].
In contrast to HSV-1, HSV-2 appears to encode yet another glycoprotein, termed gF [30,31,35,40]. Although HSV-2 gF had an electrophoretic mobility which was much faster than HSV-1 gC, initial mapping studies with recombinant viruses revealed that this protein was encoded by a region of the HSV-2 genome which was approximately colinear with the gene for HSV-1 gC (35, 40). Subsequent studies with finer structural mapping revealed a much closer collinearity between the HSV-1 gC and the HSV-2 gF coding regions [41]. In addition, it has been recently demonstrated that a monoclonal antibody against HSV-2 gF cross-reacts weakly with HSV-1 gC [42] and that a polyclonal antiserum made against HSV-1 virion envelope proteins precipitates gF [40], suggesting a possible structural homology between the two glycoproteins. Thus, it appeared that a possible homolog to HSV-1 gC was the HSV-2 gF protein. The most conclusive proof of relatedness between two proteins is to demonstrate homology at the amino acid level.
In this research we used Bioinformatics to study homology between the Herpes Simplex Virus type 1 glycoprotein C gene and Herpes Simplex Virus type 2 glycoprotein F gene by using dry lab biometric analysis of biological data using software such as (BLAST, FASTA, PDB and PHYRE2).
Materials and Methods
GenBank
GenBank ® is the NIH genetic sequence database, an annotated collection of all publicly available DNA sequences (Nucleic Acids Research, 2013 Jan;41(D1): D36-42). GenBank is part of the International Nucleotide Sequence Database Collaboration, which comprises the DNA Databank of Japan (DDBJ), the European Nucleotide Archive (ENA), and GenBank at NCBI. These three organizations exchange data daily
A GenBank release occurs every two months and is available from the ftp site. The release notes for the current version of GenBank provide detailed information about the release and notifications of upcoming changes to GenBank. Release notes for previous GenBank releases are also available. GenBank growth statistics for both the traditional GenBank divisions and the WGS division are available from each release.
We used the gene bank to detect the DNA sequences of both glycoprotein C and glycoprotein F of herpes simplex virus type 1 and type 2 respectively (https://www.ncbi.nlm.nih.gov/genbank/).
Blast
BLAST finds regions of similarity between biological sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance.
The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify members of gene families.
We used the blast programmed to detect the homology between herpes simplex virus type 1 glycoprotein C and herpes simplex virus type 2 glycoprotein F and estimate the percentage similarity as well as the homology between some other strains (https://blast. ncbi.nlm.nih.gov/Blast.cgi)
Protein Data Bank PDB
The protein data base is used to detect the amino acid sequences of both glycoprotein C and F and detect the homology between the 2 sequences (https://www.ncbi.nlm.nih.gov/pdb).
Phyre2 to analyze protein structure
Phyre2 is a suite of tools available on the web to predict and analyze protein structure, function and mutations. The focus of Phyre2 is to provide biologists with a simple and intuitive interface to state-of-the-art protein bioinformatics tools. Phyre2 replaces Phyre, in this updated protocol, we describe Phyre2, which uses advanced remote homology detection methods to build 3D models, predict ligand binding sites and analyze the effect of amino acid variants (e.g., nonsynonymous SNPs (nsSNPs) for a user’s protein sequence. This protocol will guide users from submitting a protein sequence to interpreting the secondary and tertiary structure of their models, their domain composition and model quality. A range of additional available tools is described to find a protein structure in a genome, to submit large number of sequences at once and to automatically run weekly searches for proteins that are difficult to model. A typical structure prediction will be returned between 30 min and 2 h after submission (http://www.sbg.bio.ic.ac.uk/ phyre2).
Results
Detection of DNA sequence of glycoprotein C and F Gene (Table 1)
Homology between Herpes simplex virus type 1 glycoprotein C gene and Herpes simplex virus type 2 glycoprotein F sequence ((Table 2)
The above result indicates 81% homology between Herpes simplex virus type 1 glycoprotein C gene and Herpes simplex virus type 2 glycoprotein F sequence.

Table 2: Homology between Herpes simplex virus type 1 glycoprotein C gene and Herpes simplex virus type 2 glycoprotein F sequence
Detection of Amino acid sequences of Glycoprotein C and F
The amino acid sequences of both Glycoprotein C and F were detected by Protein data bank PDB as indicated below
Herpes simplex virus type 1 glycoprotein C
translation=”MAPGRVGLAVVLWSLLWLGAGVSGGSETASTGPT
ITAGAVTNASEAPTSGSPGSAASPEVTPTSTPNPNNVTQNKTTP
TEPASPPTTPKPTSTPKSPPTSTPDPKPKNNTTPAKSGRPTKPP
GPVWCDRRDPLARYGSRVQIRCRFRNSTRMEFRLQIWRYSMGP
SPPIAPAPDLEEVLTNITAPPGGLLVYDSAPNLTDPHVLWAEGAG
PGADPPLYSVTGPLPTQRLIIGEVTPATQGMYYLAWGRMDSPHE
YGTWVRVRMFRPPSLTLQPHAVMEGQPFKATCTADAYYPRNPV
ELVWFEDDRQVFNPGQIDTQTHEHPDGFTTVSTVTSEAVGGQVP
PRTFTCQMTWHRDSVTFSRRNATGLALVLPRPTITMEFGVRHVV
CTAGCVPEGVTFAWFLGDDPSPAAKSAVTAQESCDHPGLATVRST
LPISYDYSEYICRLTGYPAGIPVLEHHGSHQPPPRDPTERQVIEAIE
WVGIGIGVLAAGVLVVTAIVYVVRTSQSRQRHRR”
Herpes simplex virus type 2 glycoprotein F
/translation=” MAFRASGPAYQPLAPRPPPARARVPAVAWIGVGAI
VGAFALVAALVLVPPRSSWGLCPCDSGWQEFNAGCVAWDPTPVE
HEQAVGGCSAPATLIPRAAAKHLAALTRVQAERSSGYWWVNGDGI
RTCLRLVDSVSGIDEFCEEL”MALGRVGLTVGLWGLLWVGVVVVLA
NASPG RTITVGPRGNASNAAPSVPRNRSAPRTTPTPPQPRKATK
SKASTAKPAPPPKTGPPKTSSEPVRCNRHDPLARYGS
RVQIRCRFPNSTRTESRLQIWRYATATDAEIGTAPSLEEVMVNVSA
PPGGQLVYDSAPNRTDPHVIWAEGAGPGASPRLYSVVGPLGRQRL
IIEELTLETQGMYYWVWGRTDRPSAYGTWVRVRVFRPPSLTIHPH
AVLEGQPFKATCTAATYYPGNRAEFVWFEDGRRVFDPAQIHTQT
QENPDGFSTVSTVTSAAVGGQGPPRTFTCQLTWHRDSVSFSRRN
ASGTASVLPRPTITMEFTGDHAVCTAGCVPEGVTFAWFLGDDSS
PAEKVAVASQTSCGRPGTATIRSTLPVSYEQTEYICRLAGYPDGIPV
LEHHGSHQPPPRDPTERQVIRAVEGAGIGVAVLVAVVLAGTAVVYL
THASSVRYRRLR
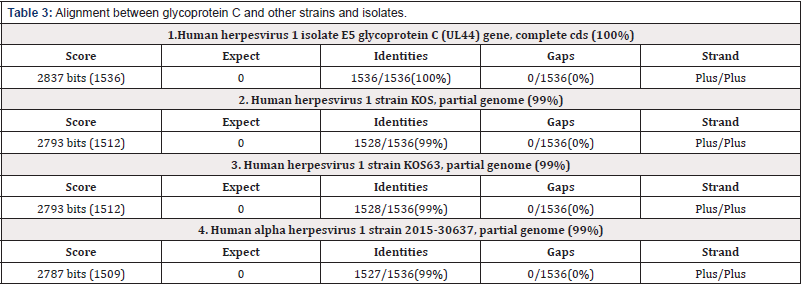
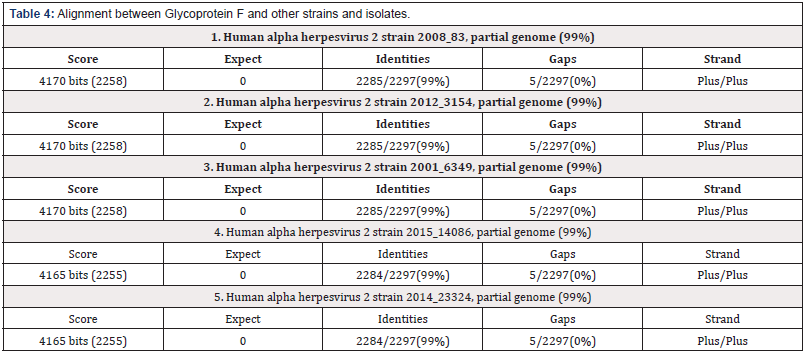
Alignment between glycoprotein C and other strains and isolates
The alignment between glycoprotein C and F of Herpes simplex virus type 1 and 2 respectively and some other steins and isolates indicates around (99-100%) homology.
Prediction of the structure of Glycoprotein C and F (Table 3) & (Table 4)
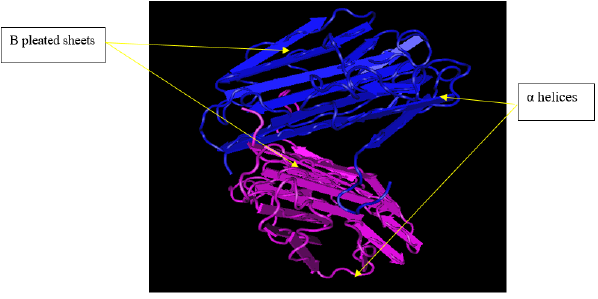
Structure of glycoprotein C: The below structure of glycoprotein C presented by PDB indicates tertiary (three dimentional) structure that harbors both α helices and β pleated sheets (Figure 1).
Structure of glycoprotein F: The above structure of glycoprotein F presented by PDB indicates tertiary (three dimensional) structure that harbors both α helices and β pleated sheets (Figure 2).
Phyre2 to analyze protein structure
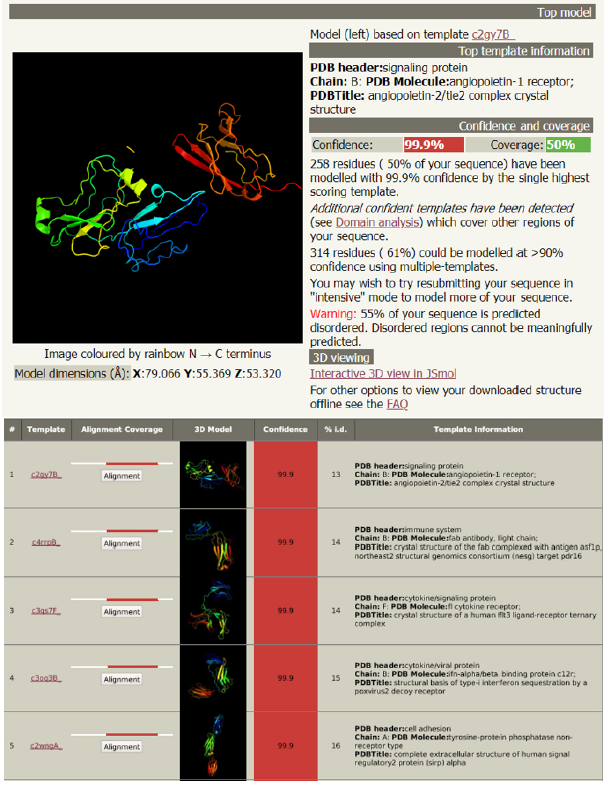
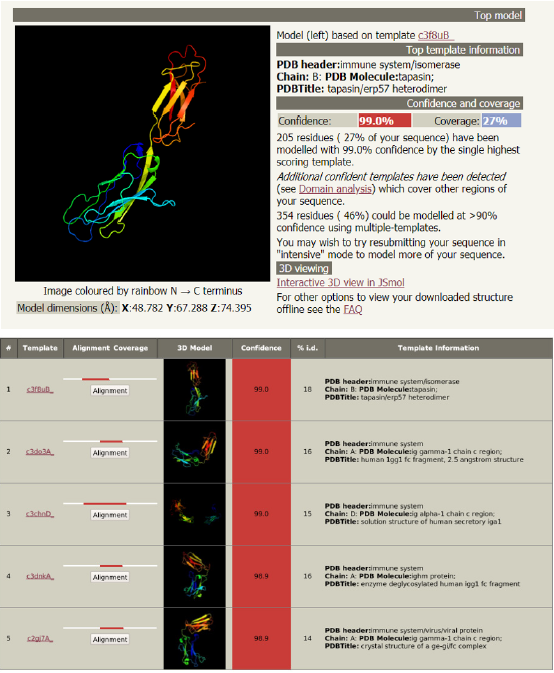
Analysis of the structure of glycoprotein C and its functions (Figure 3): Phyre2 is a suite of tools available on the web to predict and analyze protein structure, function and mutations. It showed the followings:
a. The complex crystal structure of glycoprotein C
b. The antigenic determinant site that binds with the fab region of specific immunoglobulin
c. The cytokine receptor site
d. The structural basis of interferon
e. The cell adhesion site
Phyre2 showed the followings (Figure 4):a) The heterodimer structure of glycoprotein F
b) The antigenic determinant site that binds with the fc
fragment region of specific immunoglobulinc) Solution structure of human secretory IgA
d) Enzyme deglycosylase human IgG Fc fragment
e) The complex crystal structure of glycoprotein F
Discussion
The recent flood of data from genome sequences and functional genomics has given rise to new field, bioinformatics, which combines elements of biology and computer science.
Bioinformatics is conceptualizing biology in terms of macromolecules (in the sense of physical-chemistry) and then applying “informatics” techniques (derived from disciplines such as applied math’s, computer science, and statistics) to understand and organize the information associated with these molecules, on a large-scale.
We are using Bioinformatics to study homology between the Herpes Simplex virus type 1 glycoprotein C gene and Herpes Simplex virus type 2 glycoprotein F and We found 81% homology, while [1] found the overall sequence homology between these two fragments was 68%. However, certain regions of the sequence showed either a much higher or lower degree of sequence homology than others. For example, the sequences between positions 0 and 570 of the HSV-1 and HSV-2 sequences showed only 51% homology, whereas the region between positions 570 and 1740 showed a much higher degree of sequence homology (80%). An additional highly homologous region (70%) was also found at the end of the two sequences from position 1975 to position 2419. In addition to the nucleotide sequence changes, the two genomes showed various deletions or insertions when compared with each other. The most notable was an 81-base-pairregion found at positions 346 to 426 of the HSV-1 gC sequence which is missing from the HSV-2 genome. From this overall sequence comparison, it appeared that there was a high degree of sequence homology between the HSV-1 region and the HSV-2 gF region sequenced here.
Our results help explain previous results which demonstrated that the HSV-2 gF and HSV-1 gC proteins were mainly type specific but did have type-common determinants [36,38,40,42]. Since several previous studies [32,36] demonstrated that these proteins induced predominantly type-specific antibodies, it is reasonable that the most antigenic regions of the proteins are found within the more divergent N-terminal sequences which follow the putative hydrophobic signal sequences. The hydrophilic nature of the divergent regions, along with their high content of potentiallinked glycosylation sites [45], suggests that these regions would be located on the surface of the protein. Exposure of these divergent sequences to the outside of the proteins maybe responsible for the generation of type-specific antibodies directed against these divergent epitopes. However, type common antibodies could likely also be generated by the more highly conserved carboxy-terminal three-fourths of the proteins, since hydrophilic regions conserved between gC and gF could be exposed to the outside of the proteins and may be, in one case, glycosylated (residues 363 to 366 of HSV-1 gC and 330 to 332 of HSV-2 gF). Thus, HSV-1 gC andHSV-2 gF share both type-specific and type-common determinants, but it appears that the type-specific determinants are more antigenic. Although an explanation of the type-specific and type common determinants of gC and gF must at this point be speculative, it is possible that the proteins have at least two functions, one of which is important for the viability of both viruses, the type-common domain, and one of which is specific for each virus type, the type-specific domain. Although the function(s) of gC and gF is at present unknown and viable gC minus mutants of HSV-1 have been isolated invitro [44], it is not clear that either gC or gF is indispensable to the viruses during in vivo infection of the human host and the establishment of latency. It is possible that at least some of the biological differences between HSV-1 and HSV-2, including prediction for site of infection and virulence, maybe due to the marked structural differences between the amino-terminal regions of gC and gF.
Conclusion
The results reported in this paper demonstrate that the HSV-1 gC and HSV-2 gF glycoproteins are highly homologous and that they encode type-common and type-specific domains.
References
- Donald J. Dowbenko, Laurence A Lasky (1984) Extensive Homology Between the Herpes Simplex Virus Type 2Glycoprotein F Gene and the Herpes Simplex Virus Type 1Glycoprotein C Gene. J Virol 52(1): 154-163.
- Shan Ling Hung, Sudha Srinivasan, et al. (1992) Structural Basis of C3b Binding by Glycoprotein C of Herpes Simplex Virus. J Virol 66(7): 4013- 4027.
- Baucke R, P Spear (1979) Membrane proteins specified by herpes simplex viruses. V. Identification of an Fc-binding glycoprotein. J Virol 32(3): 779-789.
- Bell S, M Cranage, L Borysiewicz, T Minson (1990) Induction of immunoglobulin G Fc receptors by recombinant vaccinia viruses expressing glycoproteins E and I of herpessimplex virus type 1. J Virol 64(5): 2181-2186.
- Draper KG, RH Costa, GTY Lee, P G Spear, E K Wagner (1984) Molecular basis of the glycoprotein-C-negative phenotype of herpes simplex virus type 1 macroplaque strain. J Virol 51(3): 578–585.
- Dubin G, I Frank, HM Friedman (1990) Herpes simplexvirus type 1 encodes two Fc receptors which have different binding characteristics for monomeric immunoglobulin G (IgG)and IgG complexes. J Virol 64(6): 2725-2731.
- Eisenberg RJ, M Ponce de Leon, HM Friedman, LF Fries, MM Frank, et al. (1987) Complement component C3b binds directly to purified glycoproteinC of herpes simplex virus types 1 and 2. Microb Pathog 3(6): 423-435.
- Friedman HM, GH Cohen, RJ Eisenberg, CA Seidel, DB Cines (1984) Glycoprotein C of herpes simplex virus 1acts as a receptor for the C3b complement component on infected cells. Nature (London) 309(5969): 633-635
- Ghosh Choudhury, NM Butcher, HP Ghosh (1990) Expression from cloned DNA of biologically active glycoproteinC of herpes simplex virus type 1 in mammalian cells. J Gen Virol 71(Pt 3): 689-699.
- Hanke T, FL Graham, V Lulitanand, DC Johnson (1990) Herpes simplex virus IgG Fc receptors induced using recombinant adenovirus vectors expressing glycoproteins E and I. Virology 177: 437-444.
- Herold BC, D WuDunn, N Soltys, PG Spear (1991) Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol 65(3): 1090-1098.
- Holland TC, FL Homa, SD Marlin, M Levine, J Glorioso (1984) Herpes simplex virus type 1 glycoprotein C-negative mutants exhibit multiple phenotypes, including secretion of truncated glycoproteins. J Virol 52(2): 566-574
- Hourcade D, VM Holers, JP Atkinson (1989) The regulators of complement activation (RCA) gene cluster. Adv Immunol 45:381-416.
- Huemer HP, M Broker, C Larcher, JD Lambris, MP Dierich (1989) The central segment of herpes simplexvirus type 1 glycoprotein C (gC) is not involved in C3b binding: demonstration by using monoclonal antibodies and recombinant. gC expressed in Escherichia coli. J Gen Virol 70: 1571- 1578.
- Johnson DC, V Feenstra (1987) Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol 61(7): 2208-2216.
- Johnson DC, MC Frame, MW Ligas, AM Cross, ND Stow (1988) Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol 62(4): 1347-1354.
- Johnson DC, MR McDermott, C Chrisp, JC Glorioso (1986) Pathogenicity in mice of herpes simplex virus type 2 mutants unable to express glycoprotein C J Viral 58: 36-42.
- Klickstein, LB, WW Wong, JA Smith, JH Weis, JG Wilson et al. (1987) Demonstration of long homologous repeating domains that are composed of the short consensus repeats characteristic of C3/C4 binding proteins. J Exp Med 165(4): 1095-1112.
- Krych M, D Hourcade, JP Atkinson (1991) Sites within the complement C3b/C4b receptor important for the specificity of ligand binding. Proc Natl Acad Sci USA 88(10): 4353-4357.
- Kubota Y, TA Gaither, J Cason, JJ O’Shea, TJ Lawley (1987) Characterization of the C3 receptor induced by herpes simplex virus type 1 infection of human epidermal, endothelial, and A431 cells. J Immunol 138(4): 1137- 1142.
- McNearney TA, C Odell, VM Holers, PG Spear, JP Atkinson (1987) Herpes simplex virus glycoproteins gC-1 and gC-2 bind to the third component of complement and provide protection against complement-mediated neutralization of viral infectivity. J Exp Med 166(5): 1525-1535.
- Para MF, RB Baucke, PG Spear (1980) Immunoglobulin G (Fc)-binding receptors on virions of herpes simplex virus type 1 and transfer of these receptors to the cell surface by infection. J Viral 34(2): 512-520.
- Perkins SJ, PI Haris, RB Sim, D Chapman (1988) A study of the structure of human complement component factor H by Fourier transform infrared spectroscopy and secondary structure averaging methods. Biochemistry 27(11): 4004-4012.
- Reid KB, DR Bentley, RD Campbell, LP Chyng, RB Sim, et al. (1986) Complement system proteins which interact with C3b or C4b. Immunol Today 7(7-8): 230-234
- Sears AE, BS McGwire, B Roizman (1991) Infection of polarized MDCK cells with herpes simplex virus 1: two asymmetrically distributed cell receptors interact with different viral proteins. Proc Natl Acad Sci USA 88(12): 5087-5091.
- Seidel-Dugan C, M Ponce de Leon, HM Friedman, RJ Eisenberg, GH Cohen (1990) Identification of C3b-bindingregions on herpes simplex virus type 2 glycoprotein C. J Viral 64(5): 1897-1906.
- Seidel-Dugan C, M Ponce de Leon, HM Friedman, LF Fries, MM Frank et al. (1988) C3breceptor activity on transfected cells expressing glycoprotein C of herpes simplex virus types 1 and 2. J Virol 62: 4027-4036.
- Tal-Singer R, C Seidel-Dugan, L Fries, HP Huemer, RJ Eisenberg et al. (1991) Herpes simplex virus glycoprotein C is a receptor for complement component iC3b. J Infect Dis 164(4): 750-753.
- Zezulak KM, PG Spear (1984) Mapping of the structural gene for the herpes simplex virus type 2 counterpart of herpes simplex virus type 1 glycoprotein C and identification ofa type 2 mutant which does not express this glycoprotein. J Viral 49(30029:741-747.
- Balachandran N, D Harnish, RA Killington, S Bacchetti, WE Rawls (1981) Monoclonal antibodies to two glycoproteins of herpes simplex virus type 2. J Virol 39(2): 438-446.
- Balachandran N, D Harnish, WE Rawls, S Bacchetti (1982) Glycoproteins of herpes simplex virus type 2 as defined by monoclonal antibodies. J Virol 44: 344-355.
- Eberle R, RJ Courtney (1981) Assay for type-specific and type-common antibodies to herpes simplex virus types 1and 2 in human sera. Infect Immun 31(3): 1062-1070.
- Eisenberg RJ, M Ponce de Leon, L Pereira, D Long, GH Cohen (1982) Purification of glycoprotein gD of herpes simplex virus types 1 and 2 by use of monoclonal antibody. J Virol 41(3): 1099-1104.
- Norrild B, H Ludwig, R Rott (1978) Identification of a common antigen of herpes simplex virus, bovine herpes mammillitis virus, and B virus. J Virol 26(3): 712-717.
- Para MF, L Goldstein, PG Spear (1982) Similarities and differences in the Fc-binding glycoprotein (gE) of herpes simplex virus types 1 and 2 and tentative mapping of the viral gene for this glycoprotein. J Virol 41(1): 137-144.
- Pereira L, D Dondero, B Norrild, B Roizman (1981) Differential immunologic reactivity and processing of glycoproteins A and gB of herpes simplex virus types 1 and 2 made in Vero and HEP-2 cells. Proc Natl Acad Sci USA 78(8): 5202-5206.
- Pereira LT, Klassen T, JR Baringer (1980) Type-common and typespecific monoclonal antibody to herpes simplex virus type 1. Infect Immune 29(2): 724-732.
- Powell K, A Buchan, C Sim, D Watson (1974) Type specific protein in herpes simplex virus envelope reacts with neutralizing antibody. Nature (London) 249(455): 360-361.
- Spear P (1980) Herpes viruses. Cell membranes and viral envelopes. HA Blough, JM Tiffaney (ed) Academic Press Inc London 2: 709-750.
- Zezulak KM, PG Spear (1983) Characterization of a herpes simplex virus type 2 75,000-molecular-weight glycoprotein antigenically related to herpes simplex virus type 1 glycoprotein C. J Virol 47(3): 553-562.
- Zezulak KM, PG Spear (1984) Mapping of the structural gene for the herpes simplex virus type 2 counterpart of herpes simplex virus type 1 glycoprotein C and identification of a type 2mutant which does not express this glycoprotein. J Virol 49(3): 741-747.
- Zweig M, SD Showalter, SV Bladen, CJ Heilman Jr, B Hampar (1983) Herpes simplex virus type 2 glycoprotein F and type 1 glycoprotein gC have related antigenic determinants. J Virol 47(1): 185-192.
- Homa FL, DJ Purifoy, JC Glorioso, M Levine (1986) Molecular basis of the glycoprotein C-negative phenotypes of herpes simplex virus type 1 mutants selected with a virus-neutralizing monoclonal antibody. J Virol 58(2): 281-289.
- Cassai E, R Manservigi, A Corallini, M Terni (1975) Plaque dissociation of herpes simplex viruses: biochemical and biological characters of viral variants. Intervirology 6(4-5): 212-223.
- Hubbard S, R Ivatt (1981) Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem 50: 555-583.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.