Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Human Identification by Amelogenin Test in Libyans
*Corresponding author: Samir Elmrghni, Faculty of Medicine, Department of Forensic Medicine and Toxicology, University of Benghazi-Libya, Benghazi, Libya.
Received: May 25, 2019; Published: July 11, 2019
DOI: 10.34297/AJBSR.2019.03.000737
Abstract
Sex typing is essential in medical diagnosis of sex-linked disease and forensic science. Gender for criminal evidence of offender is usually as the initial information for investigation. For individualization, identification of gender is performed in addition to the STR markers recently. We reported the anomalous amelogenin results of 2 male samples (out of 238 males) represented as females (Y deletions) and another 2 samples with (X deletions) in Benghazi (Libya). The frequency in both was about 0.8%. Higher than those of the other populations reported. To confirm amelogenin results of the controversial samples, DNA was further used in SRY and Y-STR typing. All samples typed as males but two showed with X chromosomes. From the results, it was highly suggested that for the controversial cases of human gender identification with amelogenin tests, amplification of SRY gene or/and Y-STR markers will be adopted to confirm the gender [1].
Keywords: Amelogenin; Libyans
Introduction
Amelogenin testing is a part of many genetic profiling multiplex systems used to determine if the sample being tested is of male or female origin 1. The gender of an offender determined from criminal evidence is commonly essential initial information for the investigation. For individualization, identification of gender is performed in addition to the STR markers. The most popular gene used in the commercial individualization kits for human gender identification is the amelogenin gene. Human amelogenin gene was first isolated and sequenced [2]. The human amelogenin gene (AMG) was precisely mapped in the p22 region on the X chromosome and the homolog (AMGL) on the Y chromosome. The location of the human amelogenin gene on maps of the X and Y chromosomes (Figure 1). Amelogenin is a protein found in developing tooth enamel and it belongs to a family of extracellular matrix (ECM) proteins. Developing enamel contains about 30% protein, and 90% of this is acerogenins (Figure 1).
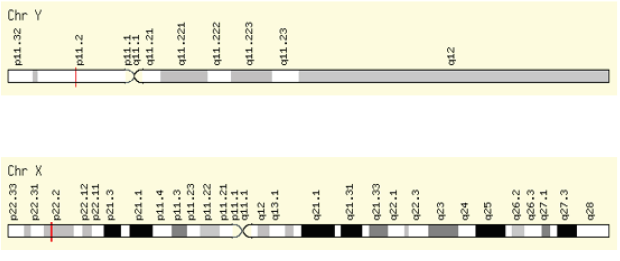
Figure 1:Genomic location of the AMELY and AMELX genes. The common position of amelogenin on the Y and X chromosomes is shown as a red line (Gene Cards Homepage 23/5/2011).
The gene for amelogenin can be used in sex determination of samples from unknown human origin through the Polymerase Chain Reaction (PCR). Using primers specific for parts of intron 1 of the gene, fragments of the gene sequence can be amplified. The X chromosome gene, AMELX, gives rise to a 106 bp amplification product (amplicon) and the Y chromosome gene, AMELY, a 112 bp amplicon hence, the AMELX contains a 6 bp deletion in intron 1. Therefore, when the amplicons are run on an agarose gel, samples from male sources (XY) will show two bands on an agarose gel (one for the 106 bp fragment and one for the 112 bp fragment), while females (XX) will show only one band. This process allows for sex determination of unknown samples. However, it was reported that two males were typed as females in 1998 by PCR amplification of amelogenin [3] and other laboratories have also observed anomalous amelogenin. The accumulated data from the above listed studies suggest that they are more likely due to deletion rather than sequence variation at the primer binding site(s). In contrast it was reported that a single phenotypically normal Caucasian male out of 327 males tested failed to show an X chromosome-specific PCR product which proved to result from the polymorphism of the primer binding sites [4].
In the current study, we decided to look for variation within the amelogenin gene in the Benghazi Population and determine the frequencies of aberrant X and Y chromosomes. Structural variation is particularly prevalent on the Y chromosome. Controversial cases have been reported, not only for males but also for females, of human gender identification by the amelogenin test. The phylogenetic context, origins and implications for male amelogenin dropouts have been reported Cadenas [5].
Material and Methods
Informed consent was obtained from 238 unrelated Libyan males (Benghazi region) same sources of studies related to Autosomal and Y chromosomal STRs.
DNA Extraction
DNA was extracted from blood stains collected on FTA® cards (Whatman, Kent, UK) using FTA® Purification Reagent (Whatman) following the manufacturer’s protocol and from buccal swabs using QIA amp® DNA Blood Mini Kit (QIAGEN, Hilden, Germany). DNA was quantified using a Step OnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, USA) [6].
PCR Amplification of Amelogenin
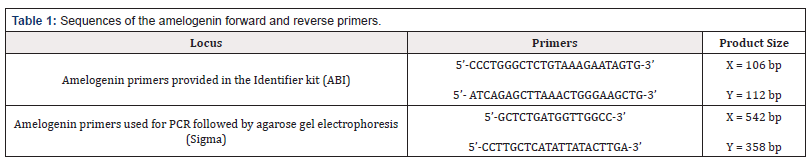
Amelogenin alleles were determined using a different primer pair from that in the Identifier Kit. The primer pair used here amplifies a different part of intron 1 of the amelogenin gene (X: p22.1-22.3 and Y: p11.2). The two primer pairs used for capillary electrophoresis [Identifier] and agarose gel electrophoresis are given in Table 1. Discs of approximately 1.2 mm cut from FTA® cards (>5 ng DNA) or 1 ng of DNA purified from buccal swabs were used to supply the target for amplification with amelogenin primers. PCR reactions were carried out in 25 mL volumes. Each PCR reaction mixture contained of 0.1 mg/ml bovine serum albumin (BSA), 12.5 l Taq DNA Polymerase Master Mix 2x (Qiagen), amelogenin forward and reverse primers (Sigma) at 0.5 M, <1 g of template, nucleasefree water to a final volume of 25 ml. The PCR reaction conditions were the same as those used to amplify part of the amelogenin gene with the Identifier kit. These were found to give good amplification with this primer pair as well [7].
PCR was carried out in a Thermocycler 9700 from Applied Biosystems. PCR conditions were as follows: starting temperature was 95°C for 11 min, (denaturing temperature 95°C for 1min, annealing 59°C for 1min, extension 60°C for 1min, 27 cycles), then 60°C for 80 minutes and held at 4°C. PCR amplification blanks (DNA replaced by equal volume of extraction reagent blank control) were used as negative controls. The PCR products were analyzed by 1.5% agarose gel electrophoresis at 120V for 1 hr. and then stained with ethidium bromide for half an hour and visualized under UV light.
Typing
Amplified products were separated and detected using the ABI Prism 310xl Genetic Analyzer (Applied Biosystems) according to the manufacturer’s recommended protocol. The data were analyzed using Gene Mapper ID v3.2 (Applied Biosystems). Alleles were assigned according to the International Society of Forensic Genetics (ISFG) guidelines for forensic YSTR [9]
Quality control
The laboratory has participated in the Y-STR Haplotyping Quality Assurance Exercise (Certified at 2010-5-20). The data were submitted to YHRD (www.yhrd.org)and received the accession number: YA003680.
PCR Amplification of Amelogenin and Agarose Gel Analysis
Amelogenin allele investigations were performed using a pair of primers (for PCR amplification followed by agarose gel electrophoresis) that were different to those provided in the AmpFlSTR® Identifiler™ kit. The same 238 DNA samples were studied. The pair of primers used amplifies a different part of intron of the amelogenin gene (X: p22.1-22.3 and Y: p11.2). A dot matrix view showing regions of similarity between the AMELY and AMELX genes based upon BLAST (Basic Local Alighment Search Tool) results is shown in Figure 2. The sequences of primers used for capillary electrophoresis (included in the AmpFlSTR® Identifiler™ kit) and the primers used for PCR amplification and agarose gel electrophoresis in this section are given in Table 1 (Figure 2).
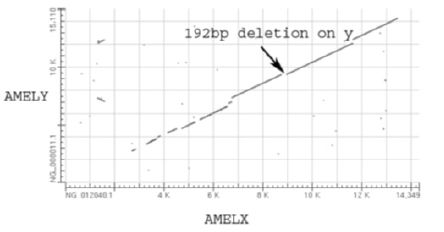
Figure 2: Dot matrix view showing regions of similarity between the AMELY and AMELX genes based upon BLAST (Basic Local Alighment Search Tool) results. The lines shown in the plot represent the alignments found by BLAST (blast.ncbi.nlm.nih.gov) (the number of lines = the number of alignments found by BLAST). The 192 bp deletion on the Y chromosome (incorporated into the amplicon obtained using our modified primers) is observed, however, the 6 bp deletion on the X chromosome (incorporated into the amplicon obtained using the AmpFlSTR® Identifiler™ kit) is too small to be resolved at this scale. The nucleotide base pair numbers observed on the X and Y axes are equivalent to those observed in the DNA sequences.
Discs of approximately 1.2 mm cut from FTA® cards (>5 ng DNA) (following washes with Whatman FTA Purification Reagent, according to the manufacturer’s protocol) or 1 ng of DNA purified from buccal swabs were used for PCR amplification with the amelogenin primers. PCR reactions were carried out in 25 ml volumes. Each PCR reaction mixture contained: 1 ml bovine serum albumin (BSA) (2mg/mL), 12.5 ml Taq DNA Polymerase Master Mix 2x (Qiagen), 1ml of each amelogenin primer (forward and reverse, 10M) (Sigma), 3 ml of template DNA (1 ng) or 2 FTA discs (>5 ng DNA), and nuclease-free water to a final volume of 25 ml.
The PCR reaction conditions were the same as those used to amplify the amelogenin gene with the AmpFlSTR® Identifiler™ kit. These were found to result in amplicons with the modified pair of primers as well. PCR was carried out in a Thermocycler 9700 (Applied Biosystems). PCR conditions were as follows: starting temperature of 95°C for 11 min, followed by 27 cycles of denaturing at 95°C for 1min, annealing at 59°C for 1min, and extension at 60°C for 1min, followed by 60°C for 80 minutes and holding at 4°C. PCR amplification blanks (DNA replaced by an equal volume of extraction reagent blank control) were used as negative controls. The PCR products were analysed by 1.5% agarose gel electrophoresis at 120V for 1 hr. and then stained with ethidium bromide (Sigma) for half an hour and visualized under UV light.
Results and Discussion of the amelogenin gene analysis in the Benghazi Population
Samples from the same 238 Libyan males from Benghazi, who had been typed for Y chromosomal STRs and autosomal STRs, were tested for a different region of the amelogenin gene from that included in the AmpFlSTR® Identifiler™ kit. This was in order to see whether there were variations within the amelogenin gene in our Benghazi population sample regarding abberations on the X and Y chromosomes Of the 238 unrelated male samples analysed using the amelogenin primers, 2 samples (0.008%) were identified as being female (Y null) (Figure 3 & 4) and the rest as male (Figure 3-6).
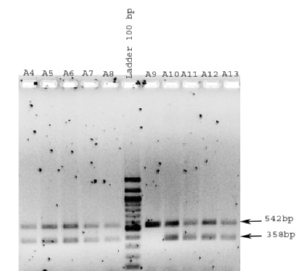
Figure 3: Benghazi male DNA samples were PCR amplified using primers for the amelogenin gene and analyzed by agarose gel electrophoresis. Two bands (representing the X and Y chromosomes) are observed for samples A4-A8 and A10-A13, whilst only one band is observed for sample A9 (known to be male but shown as female).

Figure 4: Benghazi male DNA samples were PCR amplified using primers for the amelogenin gene and analyzed by agarose gel electrophoresis. Two bands (representing the X and Y chromosomes) are observed for samples A14 and A16, whilst only one band is observed for sample A15 (known to be male but shown as female).
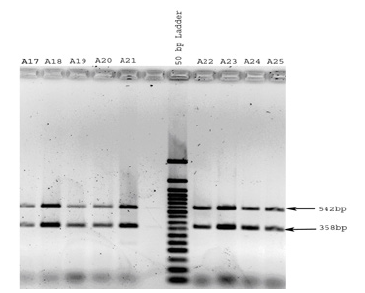
Figure 5: Benghazi male DNA samples were PCR amplified using primers for the amelogenin gene and analyzed by agarose gel electrophoresis. Two bands (representing the X and Y chromosomes) are observed for all samples (A17-A25).
For example, Samples A9 and A15 (Figures 3 & 4) only generated the X chromosomal amelogenin amplicon, so typed female, although the sample was from a male individual with a full Y-STR haplotype [7] and a male result using the AmpFlSTR® Identifiler™ kit which includes primers for amplification of the amelogenin gene [8]. This suggests that the Y chromosome is largely intact and that the mutation or deletion preventing PCR amplification of the Y amelogenin gene is confined to a limited region beyond that tested in the AmpFlSTR® Identifiler™ kit and does not affect any Y-STR loci. The two Y null males are unrelated and have different surnames (Figure 5).
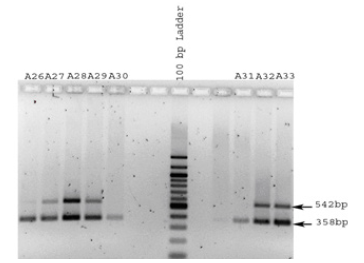
Figure 6:Benghazi male DNA samples were PCR amplified using primers for the amelogenin gene and analyzed by agarose gel electrophoresis. Two bands (representing the X and Y chromosomes) are observed for samples A26-A29 and A32 and A33. Samples A30 and A31 revealed an X abberation or deformity (a Y band only on the agarose gel and band 542 bp (representing the X chromosome) missing). This may be due to a mutation (the test was repeated twice).
The frequency of Y null in this Benghazi population (0.008%) was different from other studies depending on the amelogenin gene testing method used. In an Austrian study, among the 29,432 phenotypic male individuals stored in the Austrian National DNA database, 6 individuals (0.018%) lack the amelogenin Y-specific PCR product which was confirmed using alternative amelogenin primers [9]. These samples were confirmed to be from males by amplification of Y-STR and SRY (sex-determining region of Y chromosome) genes. A high frequency of deletion for the Y amelogenin region in a male Indian population group has been documented [10]. In one study, using the AmpFlSTR Profiler Plus kit, amelogenin tests have shown a significant proportion of Y null alleles in an Indian ethnic group (3.6% frequency) and Malay ethnic group (0.88% frequency) [9,11]. (Figure 6-8). The possibility was reported that partial Y deletion forms a relatively old and stable haplotype in the Indian ethnic group [11]. In the Indian population group the high Y null frequency was due to deletion of the fragment Y(p)11.2 whilst Steinlechner [9] PCR amplified a fragment of the SRY gene in Austrians. In another study, the frequency of deletion for the Y amelogenin region in a male Indian population group was 0.23% (10 individuals out of 4,257 male samples analyzed from 104 different endogamous populations of India) and deletion of the fragment Y (p) 11.2 was detected, so the male samples would have been identified as females in such cases [12].
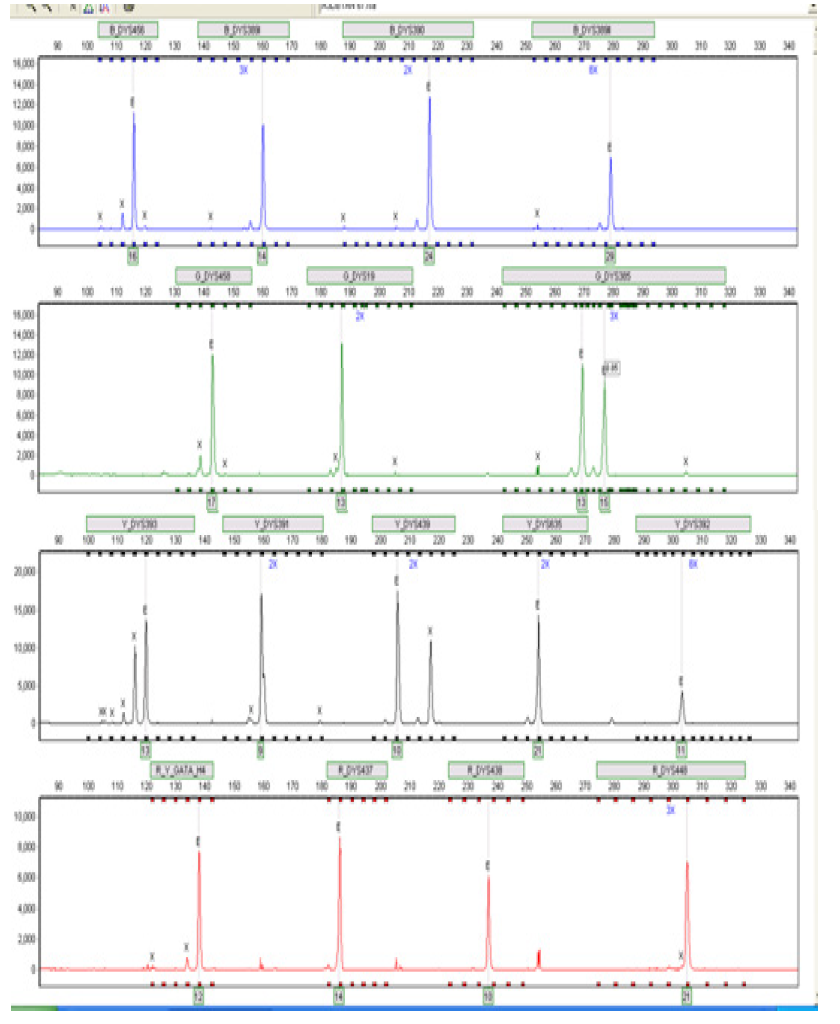
Figure 7:Electropherogram showing the 17 Y-STR loci detected in male A9 using the AmpFlSTR® Y filer kit. This individual typed female in the agarose gel electrophoresis-based test shown in Figure 6.4 but male when typed using the AmpFlSTR® Y filer kit (which is specific for males).
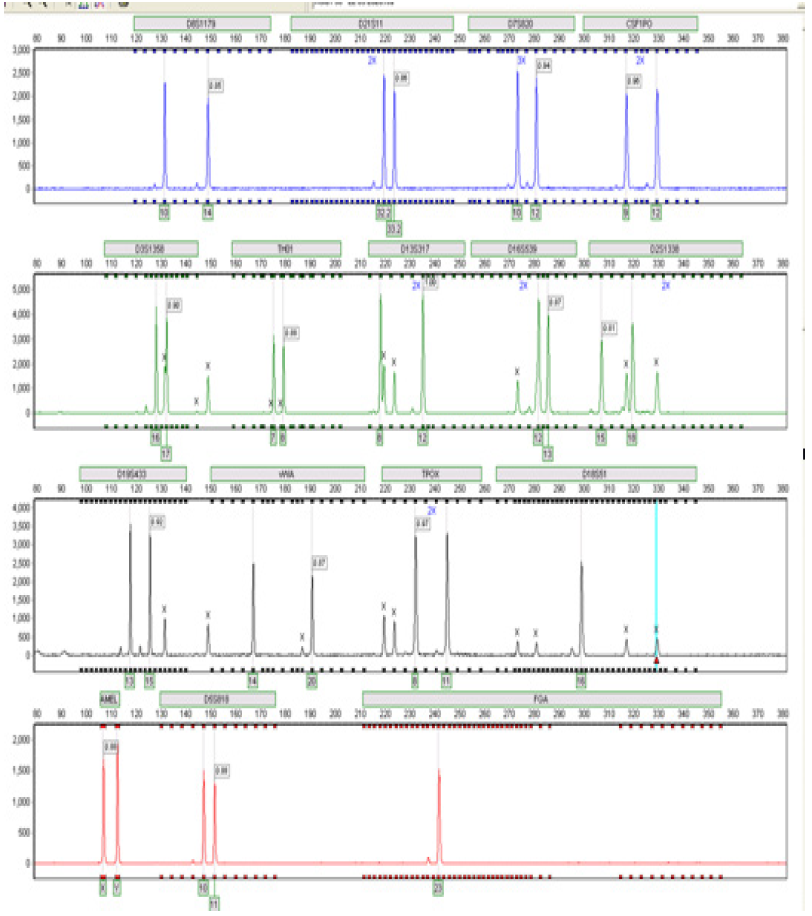
Figure 8:Electropherogram showing the 15autosomal loci detected in male A9 using the AmpFlSTR® Identifiler kit. This individual typed female in the agarose gel electrophoresis-based test shown in Figure 6.4 but an XY amelogenin gene result was obtained using the AmpFlSTR® Identifiler kit.
X deletions or aberrations were seen in two male samples from the Benghazi population of the current study, resulting in a frequency of 0.008% (2 samples of 238). These individuals, who are male, were represented as male when typed using the AmpFlSTR® Y filer and AmpFlSTR® Identifiler kits in the current study, and for the same reason a Y deletion occurred in the previous example, X deletions may arise. In a study on a Taiwanese population, DNA samples were PCR amplified using the Quantifiler Y Human Male DNA Quantification Kit and the AmpFISTR Yfiler PCR Amplification Kit to test for the SRY gene and Y-STRs, respectively. The frequency of deletions was about 0.00625% for males (5 samples of 80,000) and 0.015% for females (3 samples of 20,000), and lower than those reported in other populations [13].
References
- Sullivan KM, Mannucci A, Kimpton CP, Gill P (1993) A rapid and quantitative DNA sex test: fluorescence- based PCR analysis of X-Y homologous gene amelogenin. BioTechniques 15(4): 636-641.
- Shimokawan H, Tamura H, Ibaraki K, Sasaki S (1989) Human amelogenin gene In Tooth Enamel V Florence Pub Yokohama 301-305.
- Santos FR, Pandya A, Tyler Smith C (1998) Reliability of DNA-based sex test. Nat Genet 18(2): 103.
- Shadrach B, Commane M, Hren C, Warshawsky I (2004) A rare mutation in the primer binding region of the amelogenin gene can interfere with gender identification. J Mol Diagn 6(4): 401-405.
- Cadenas AM, Regueiro M, Gayden T, Singh N, Zhivotovsky LA, et al. (2007) Male amelogenin dropouts: phylogenetic context, origins and implications. Forensic Sci Int 166(2-3): 155-163.
- Gusmão JM, Butler A, Carracedo P, Morling M, Prinz L, et al. (2006) DNA Commission of the International Society of Forensic Genetics, DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis, Forensic Sci Int 157(2-3): 187-197.
- Elmrghni YM, Coulson-Thomas, M Kaddura, RA Dixon, DR Williams (2012) Population genetic data for 17 Y STR markers from Benghazi (East Libya). Forensic Sci Int Genet 6(2):224-227
- Elmrghni, S, Yvette M Coulson-Thomas, Mahmoud Kaddura, Ron A Dixon, DR Williams (2011) Genetic data provided by 15 autosomal STR loci in the Libyan population living in Benghazi. Forensic Sci Int Genet 6(3): e93-e94.
- Steinlechner M, Berger B, Niederstatter H, Parson W (2002) Rare failure in the amelogenin sex test. Int J Legal Med 116(2): 117-120.
- Thangaraj K, Reddy A, Singh L (2002) Is the amelogenin gene reliable for gender identification in forensic casework and prenatal diagnosis? Int J Legal Med 116(2): 121-123.
- Chang YM, Burgoyne LA, Both K (2003) Higher failures of amelogenin sex test in an Indian population group. J Forensic Sci 48(6): 1309-1313.
- Chang YM, Perumal R, Keat PY, Yong RY, Kuehn DL, et al. (2007) A distinct Y-STR haplotype for Amelogenin negative males characterized by a large Y(p)11.2 (DYS548-MSY1-AMEL-Y) deletion. Forensic Sci Int 166(2-3): 115-120.
- Kao MS, Li-Chin Tsai, James Chun-I Lee, Hsing-Mei Hsieh (2007) Controversial cases of human gender identification by amelogenin test. Forensic Science Journal 6(2): 69-7



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.