Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Modern Era of Rice (Oryza Sativa L.) Genomics for Precise Genomics-Assisted Drought Breeding
*Corresponding author: Muhammad Mahran Aslam, Department of Molecular and Cellular Biology, Guelph University of Canada, Canada.
Received: May 22, 2019; Published: June 13, 2019
DOI: 10.34297/AJBSR.2019.03.000680
Abstract
Rice (Oryza sativa L.) is an important food crop and requires larger amount of water throughout its life cycle as compared to other crops. Hence, water related stress cause severe threat to rice production. Drought is a major challenge limiting rice production. The screening of rice germplasm under drought and its characterization at the morphological, genetic, and molecular levels revealed the existence of genetic variation for drought tolerance within the rice gene pool. The improvements made in managed drought screening and selection for grain yield under drought have significantly contributed to progress in drought breeding programs. The availability of rice genome sequence information, genome-wide molecular markers, and low-cost genotyping platforms now makes it. Drought possible to routinely apply marker-assisted breeding approaches to improve grain yield under drought. Grain yield Grain yield QTLs with a large and consistent effect under drought have been identified and successfully Genetics pyramided in popular rice mega-varieties. The transgenic approach Proteomics is successful in generating drought tolerance in rice under controlled conditions, but field-level testing is necessary. Genomics-assisted drought breeding approaches hold great promise, but a well-planned integration Allele mining with standardized phenotyping is highly essential to exploit their full potential.
Keywords: Genomics; Proteomics; Transcriptomics; Drought; Rice and Genetics
Introduction
Water is an important factor in agricultural and food production, yet it is a highly limited resource (Wang et al, 2012). Water deficit stress causes extensive loss to agricultural production worldwide, thus being a severe threat to sustainable agriculture. Feeding continuously increasing population with depleting water supply requires crop varieties that are highly adapted to dry environments [1]. Rice plays a major role as a staple food, supporting more than three billion people and comprising 50% to 80% of their daily calorie intake [2]. Drought stress severely impairs its production. Worldwide, drought affects approximately 23 million hectares of rainfed rice [3].
Rice is highly susceptible to drought stress throughout its life cycle, but huge economic losses or even complete crop failures are observed if stress occurs during flowering [4]. The capacity of the rice plant to sustain itself and to reproduce in limited water conditions is crucial for rice production in years of drought [5,6]. Thus, it is imperative for rice breeders to develop droughttolerant high-yielding rice cultivars. Even though several efforts were made to breed for drought tolerance by including tolerant donors in breeding programs, there are few examples of improved rice cultivars that combine acceptable yield potential and drought tolerance. This is mainly because of the genetic complexity of drought tolerance due to its polygenic inheritance, low to medium heritability, significant genotype and environmental interactions, and the confounding effects of other abiotic stresses on drought [7-9]. In view of this, breeders have to screen and select a very large number of genotypes over seasons, years, and locations to successfully develop drought-tolerant varieties. The whole process is time-consuming, labor-intensive, and expensive. Drought breeding efforts over the past two decades have been successful in standardizing protocols for managed drought screening infield conditions by selecting sites having climatic conditions and soil types similar to those of the drought-prone target environments. Trait phenotyping with grain yield under drought has been proven as an efficient criterion and is currently being used in drought breeding programs [10].
Rice is the first food crop with both of its subspecies, indica and japonica, sequenced [11,12], and also ten wild species of rice belonging to different genomes are being sequenced (www.map. org). Rice has an enormous wealth of genomics and bioinformatics resources that can be used to speed up the breeding process. Recent advances in genomics technologies such as molecular markers and genetic engineering have made rapid strides in the understanding of the molecular basis of drought tolerance and enabled the identification of genes/QTLs for yield and yield-related traits under drought. Now, the stage is set for effective integration of genomics technologies with breeding activities to improve drought tolerance in rice. In our review, we discuss the recent advances in understanding the molecular basis of drought tolerance and the application of genomics technologies in improving drought tolerance in rice.
Genetics and Genomic Prospective of Rice in the Era of Drought Tolerance
Genetic prospective of rice
The knowledge of genetic basis of drought tolerance is vital approach to start a breeding program. It will be more beneficial to evaluate a crop variety under any adverse environmental condition. Genetic of drought is controlled by polygenes [13]. Genetic study of physical and yield governing traits as well as root traits showed positive interaction with environment followed by polygenic inheritance like additive and dominant type of gene action [13,14-17]. Positive Epistatic gene action were also observed [18,19]. It is estimated that secondary characters of rice cultivar may be beneficial to combat drought tolerance but in field this assumption was not observed yet. Medium to high heritability was observed for grain yield under drought tress breeding on this criteria will be proved enhance rice yield under drought.
Genomics prospective of rice
Advances in molecular genetics and genomics rice genome is completely sequenced and it is now possible to develop a diverse variation from a single QTL [20,23]. There are several approaches i.e. association mapping, genome-wide selection (GWS). Linkage mapping, Marker assistant selection which can be utilized to develop a drought tolerance rice variety [24]. Genomic diversification can made with such technique’s QTL × QTL connections, QTL × environment interactions and QTL introgression. A complete set of these variation is key factor to initiate marking assistant program of drought tolerance in rice [25]. Variability is found in major-effect QTLs for grain yield could be utilized for MAS for drought tolerance in rice.
Rice genomics resources and tools for precise genomicsassisted drought breeding
Advances in molecular genetic lead to high value of genomic diversity, these advance techniques are rapid and very convenient in breeding point of view [26]. As well as improvement in bioinformatics and genomics approach have been proved more beneficial for biotechnologist and molecular researchers
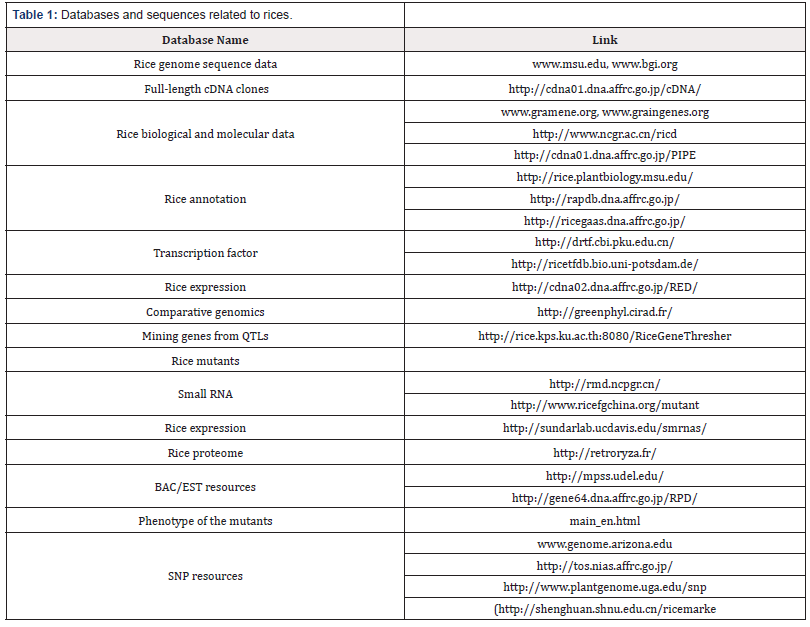
Rice is the first sequenced food crop and has been at the leading position in plant genomics because of its significance in world food 1309 B.P.M. Swamy, A. Kumar / Biotechnology Advances 31 (2013) 1308–1318 security. Rice has small genome size of 390 Mb and known as a model in cereal crops. The genome of have two sub species, japonica (cv. Nipponbare), indica (cv. 93–11), and organelle genome such as mitochondrial and chloroplast genome has been sequenced and genome of ten different wild rice are being sequenced. All this information is placed in the public domain and is easily accessible for breeding point of view (www. gramene.org, www.msu.org,www.bgi.org). BAC end sequences, Bacterial artificial chromosome (BAC) libraries, and whole-genome sequence approaches develop various marker systems and led to the construction of physical maps and great-diverse genetic maps [27,28]. Rice contain both functional and non-functional markers which include more than 20,000 SSR markers and over one million SNPs and Indels including useful and non-useful markers [29,30]. For the use of molecular markers in diversity analysis, has proved to be landmarks in molecular genetics, mapping genes/QTLs for various agronomic traits, and their use in marker-assisted breeding (MAB) and also identify candidate genes in positional cloning of QTLs for complex traits studies under various environmental stresses like drought [31]. The accessibility of rice functional genomics include cDNA libraries, transcriptome maps, expressed sequence tags (ESTs), TILLING array networks, insertion mutant genomics libraries and advance detection technique such as realtime PCR, macro array and microarray, serial analysis of genome expression (SAGE), massive parallel signature sequences, genomewide association mapping etc. [32], and different proteomics, Transcriptomic studies and metabolomics study have greatly useful for drought tolerance global and local gene expression analyses to evaluate and characterize candidate genes [33-35]. For particular gene expression we must have Transcriptomic information regulatory machinery and housekeeping gene study help us to understand the pathways of gene regulation [36-38]. Table 1.shows rice genomics resources and tools. For genetic engineering, rice crop has modified for tissue culture, direct and indirect gene cloning i.e. agrobacterium mediated and biolistic technique of gene introgression, advance detection method labelled and non-labeled assay for gene confirmation techniques to study copy number of gene at protein or mRNA level [39-41]. For developing, evaluating, and handling transgenic plants there are already international standard biosafety measures. The well-established sources of various types of molecular data and advanced bioinformatics tools have subsidized to searching, querying, depositing, and analyzing all types of information that is being explored by rice scientists [42]. Recent landmarks in rice genome enabled scientist as well as rice breeders to utilized natural and induced diversity for the improvement of Rice crop (Figure 1) [43].
Recent platform developed for Rice Genomics for drought tolerance
Meanwhile the accomplishment of whole-genome sequencing in rice, numerous well-designed genomics platforms have been recognized in the historical period [44,45]. Stages of metabolomics, proteomics, and phenomics have likewise been progressively wellknown and amended, and compatible platforms of bioinformatics investigation and catalogues have also been set up in rice.
Transcriptomics
Considerate the genes that govern response to drought stress is beneficial in breeding rice with improved drought tolerance. The response to drought stress in rice could also be an advanced development involving several functionally interconnected genes unfold throughout the order. Natural phenomenon Analysis permits the analysis of the expression of thousands of genes in an extremely extra comprehensive and holistic suggests that, providing a world image of changes occurring inside the transcriptome of varied components of the plant beneath stress conditions. Expression analysis results is accustomed establish candidate genes for specific drought tolerance-related traits or physiological mechanisms associated with stress-induced adaptive processes [38,46,47]. In rice, several efforts were created to grasp the differential expression of genes under drought stress by transcription identification pattern RT-PCR and microarray technologies [48] distributed microarray analysis of rice panicles selected from a drought stressed plant and reported up-regulation of the many stressinduced genes like GTP-binding molecule 3, sugar synthase-6, heat shock cognate molecule, deoxyribonucleic acid repair molecule, reductase, zinc finger molecule, protein depolymerizing issue, and polysaccharide esterase [49] far-famed sixty 2 drought-inducible genes in 2-week-old drought-stressed rice seedlings. Widespread rice landrace N22 (Nagina-22) could also be a natural different for lots of molecular characterization work because of its drought tolerance. A drought-induced DNA library was made of N22 and transcription identification was performed, 589 acknowledged stress-related genes were far-famed, and, pattern this knowledge, candidate genes were deduced for drought QTLs [50]. In another study pattern N22, ESTs were generated from drought-stressed rice seedlings; most of the stress-induced genes were related to primary metabolic pathways, transcription, and conjointly the interpretation methodology [51]. Differential expression analysis of upland and lowland rice cultivars victimization cDNA-AFLP unconcealed that quite eight of the entire genes clothed were upregulated by drought stress in each cultivars.
Fifty-seven and thirty-eight genes were specifically expressed in upland and lowland cultivars, severally. Among the differentially expressed genes, genes for cell rescue, drought defense, signal transduction, nucleotides, organic compound biogenesis, and genes for plant growth and development were specifically expressed in upland rice, whereas genes for macromolecule and ester degradation were specific to lowland rice cultivars [52]. The spatial temporal expression patterns of two-component system ( TCS) genes in rice underneath drought stress showed a differential expression pattern in varied organs underneath stress [53-55]. Reportable genes and signal pathways concerned within the anatomical and morphological developments of rice roots, notably for crown root initiation underneath drought stress [56]. conducted a tissue-specific organic phenomenon study in droughtstressed rice roots, during which sixty-six transcripts were known. Out of those, four transcripts were mapped inside the QTLs for root growth underneath water deficit. In another study, [57] discovered differentially expressed genes in leaves and roots of upland rice and lowland rice. Combining our data of genes concerned in growth and development, reconciling mechanisms with genes that are differentially expressed underneath drought may facilitate determine necessary target traits for drought resistance. The differentially expressed genes also can function candidate genes for additional characterization and for allele-morph mining.
Proteomics
Histone modifications, DNA methylation, and microRNAs are the 3 main epigenetic regulators of drought tolerance. Underneath drought stress, simple protein modifications like acylation and/ or methylation pattern among the promoters or writing regions of cistrons cause cistron activation or gene silencing, leading to tolerance of drought stress [58]. Pyrimidine methylation and demethylation by DNA methyltransferases are epigenetic mechanisms adopted by the upper organisms in response to worry. It causes differential regulation of organic phenomenon through either silencing or over [59,60]. There are reports demonstrating that DNA methylation-regulated organic phenomenon could be a response to drought stress. A recent study exploring the genome-wide DNA methylation standing of 2 rice cultivars (DK151 and IR64) with totally different tolerance of drought disclosed important variations within the methylation patterns between the 2 genomes [32]. Methylation and demethylation changes were elicited underneath drought conditions during a biological process and tissue-specific manner and that they accounted for twelve.1% of the entire site-specific methylation variations between the 2 lines. Notably, seventieth of the drought-induced methylation changes were reversed once recovery, and twenty nine remained dateless [32]. Changes in methylation pattern in rice cultivars were reported by [61]. These observations recommend that DNA methylation changes play a task within the response of rice to dehydration conditions in all probability by activating or deactivating stress-responsive genes and resulting in adaptation to drought conditions. The fiber non-coding RNAs like microRNAs (miRNAs) and SiRNAs are concerned within the epigenetic method of drought stress. To date, many drought-responsive microRNAs are known in plants. MicroRNAs play vital roles in post-transcriptional cistron regulation by repression of template RNA translation. In rice additionally, small polymer regulated drought response has been reported [38,62].
Metabolomics
Plants react to drought stress by accumulating various metabolic compounds such as proline, glycine-betain, pinitol, carinitine, mannitol, sorbitol, polyols, trehalose, sucrose, oligosaccharides, and fructans in large quantities [63]. These are chemically dissimilar compounds; they keep the surface of the proteins hydrated, resulting in decreased water potential and facilitating continuous water movement, which might contribute to sustaining physiological processes such as stomatal opening, photosynthesis, and expansion of cell growth. The role of metabolites in drought tolerance has been thoroughly reviewed in earlier reports [64,65].
Epigenomics
Histone modifications, DNA methylation, and microRNAs are the three main epigenetic regulators of drought tolerance. Under drought stress, histone modifications such as acetylation and/ or methylation pattern within the promoters or coding regions of genes cause gene activation or gene silencing, resulting in tolerance of drought stress [59]. Cytosine methylation and demethylation by DNA methyl-transferases are epigenetic mechanisms adopted by the higher organisms in response to stress. It causes differential regulation of gene expression through either silencing or over [60,61]. There are reports demonstrating that DNA methylationregulated gene expression is a response to drought stress. A recent study exploring the genome-wide DNA methylation status of two rice cultivars (DK151 and IR64) with different tolerance of drought revealed significant differences in the methylation patterns between the two genomes [66]. Methylation and demethylation changes were induced under drought conditions in a developmental and tissue-specific manner and they accounted for 12.1% of the total site-specific methylation differences between the two lines. Notably, 70% of the drought-induced methylation changes were reversed after recovery, and 29% remained unaltered [67]. Changes in methylation pattern in rice cultivars were reported by [32]. These observations suggest that DNA methylation changes play a role in the response of rice to dehydration conditions probably by activating or deactivating stress-responsive genes and leading to adaptation to drought conditions. The single-stranded non-coding RNAs such as microRNAs (miRNAs) and SiRNAs are involved in the epigenetic process of drought stress. To date, hundreds of droughtresponsive microRNAs have been identified in plants. MicroRNAs play important roles in post-transcriptional gene regulation by repression of mRNA translation. In rice also, micro RNA regulated drought response has been reported [38,39].
Genetic engineering for drought resistance in rice
Drought resistance in rice can be overcome by genetic engineering techniques (Hervé and Serraj, 2009; Mathur et al., 2008). Direct and indirect genetic transformation techniques (e.g. agrobacterium mediated and biolistic) were applied in rice genome for drought resistance. These techniques proved to be very efficient in past few decades. Large numbers of candidate genes were observed against drought stress which was directly involved in post transcriptional modification, signal cascades and metabolite triggering (Yang et al., 2010). Genetic engineering was found efficient technique that allows gene pyramiding for biotic and abiotic stresses. (Cattivelli et al., 2008).
Transcription analysis enables gene expression for specific environmental stress especially for drought at any chronological crop stage. Genetic expression of MAP kinase related genes were found in drought stress (Agrawal et al., 2003); Other heat related proteins like proline ,LEA and HSP and other osmolytes (Sato and Yokoya., 2008; 62; Xu et al., 1996; Zhu et al., 1998); DREB1/CBF is DREB genes (Dubouzet et al., 2003endo-1,3-glucanase (Akiyama and Pillai, 2001); NAC genes (Hu et al., 2006; Leung, 2008); ), Ca Dependent protein kinase (Saijo et al., 2000); and trehalose ( sugar compounds) (Garg et al., 2002; Jang et al., 2003). Rice genome modified with such gene pyramiding protocol would be better adapted to new environment under drought. Overall efforts interpreted that proper genetic resource and techniques would be very effective for gene expression under stress. Durability and stable performance of genetic assembly of any specie is key factor for breeder. Transcriptional factor along with specific genes have been incorporated in rice genome for durable and stable expression of protein under various type of seasonal fluctuates.
Transgenic rice germplasm has been widely evaluated invitro condition for better result it is needed to be evaluated in environmental condition. Hence transgenic approaches are preferred over traditional breeding program which is very laborious and time consuming. Genetic engineering is novel breeding technique widely used along the world to combat the global warming specially drought. It is needed that genetic engineering in rice would be very selective and appropriable method especially for drought stress (Bhatnagar-Mathur et al., 2008; Yang et al., 2010).
Water is important component for Food security yet it is scarce resource in the world [67]. Water scarcity limit the agricultural growth worldwide. It is urgent need to develop a drought tolerant variety to produce quality food for the growing population [1]. Rice is staple food along the worldwide which feed three billion population, depending upon daily dietary needs [2]. Rice crop is severely affected by water deficiency which significantly reduces per hectare production. Rainfed rice yield is reduced by drought stress approximately 23 million hectare worldwide [3].
Rice crop is highly sensitive to water scarcity but during flowering stage, it was observed that the water stress cause economic loss on large scale. [4]. The ability of rice plant to stand in water scarcity condition is important for production of rice during drought season [5,6]. Breeder must develop rice varieties suitable for water deficit conditions. Recently several research programs have been introduced to develop drought tolerance rice cultivars utilizing multidiscipline approaches. .polygenic traits were introgressed in rice germplasm for better adaptation of rice variety under adverse biotic and abiotic stresses. Thus, it is imperious for breeders to develop cultivars of high yielding and drought tolerant. [7,9]. Genetic variability and durability of drought tolerance genes are novel approach for the stable production of rice against drought. Screening of these lines and introgressed to cultivated germplasm is highly demanding objective for development of drought tolerance rice variety. But this process required time, cost and human capital. Research has been accomplished to develop drought tolerance rice variety in past few decades. Maximum production with sufficient adoptability of cultivar under drought condition is prerequisites for the initiation of breeding program [10].
Cultivated rice has two wild species named as O. indica and O. japonica, both have completely sequenced [12,13], as well as more than 10 rice wild progenitor have been sequenced (www.map.org). Breeding of rice crop can be modified by utilizing bioinformatics and genomic tools which had been widely used in rice genome. Genetic engineering and MAS proved to be latest and innovative breeding strategy to evaluated genetic architecture of drought tolerance genes in rice. By utilizing these strategies rice production would be increased under drought conditions. Yield related genes/ QTLs expressed under drought condition have been successfully identified Now, it is highly demanding objective to incorporate these genes into rice by using Genetic engineering approach. This review will benefits the scientist to develop drought tolerance rice cultivar by understanding the genetic and genomics application of innovative research tools in breeding programs.
An integrated approach is the way forward
For drought tolerance, breeding efforts in rice have clearly revealed that a multidisciplinary approach is necessary. Utilizing Standard approach to study the physiological basis of drought tolerance helps to determine the specific anatomical and morphological diversity, and pathways. Selection for donor parents above mention variation and approaches proved to be useful in molecular and traditional breeding program for drought tolerance [7]. With the help of diverse genomics technologies such as molecular markers, proteomics, Transcriptomics, epigenomics, metabolomics and, together with conventional breeding approaches facilitated to identification and characterization of genes/QTLs that is responsible for specific gene regulation under drought (Ashraf, 2010) [68]. Marker assisted breeding is proved to be very rapid and efficient technique for the introgression of drought tolerance QTLs/ genes in rice [10,69]. A transgenic method allows the manipulation of drought-tolerant genes from different sources [70-75]. Through breeding procedures after the evaluation of transgenic plants and gene incorporation, these genes can be transferred to other genetic database [76-80]. Functional genomic study may be very crucial in future crop improvement. Breeder can only utilized these technologies when reasonable variation and integration is observed in rice germplasm.
Conclusions
Drought tolerance trait is very complex genetic character which is main cause of drought tolerance rice germplasm However drought stress and its mitigation strategies were explored in last few years. Genetic variability for specific trait is key factor to start a breeding program. Genetically and physiological screening of rice germplasm for grain yield, QTLs, genomic marker and MAS would be very effective to develop drought resistant rice cultivar. There are several Quantitative trait loci for Rice yield under drought have been transformed and would be utilized in future research. Advancement in functional genomics significantly helps to evaluate various molecular pathways and genetic resource participated in drought tolerance. Rice genome can be edited for drought tolerance and genetic pyramiding of drought responsive genes would be revolutionize rice productivity. Drought tolerance variety of rice can be developed through genetic engineering techniques.
References
- Foley J A, Ramankutty N, Braumann K A, Cassidy ES, Gerber J S, et al. (2011) Solutions for a cultivated planet. Nature 478: 337-342.
- Khush GS (2005) What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol Biol 59(1): 1-6
- Serraj R, McNally KL, Slamet Loedin I, Kohli A, Haefele SM, et al. (2011) Drought resistance improvement in rice: An integrated genetic and resource management strategy. Plant Prod Sci 14(1): 1-14.
- Pantuwan G, Fukai S, Cooper M, Rajatasereekul S, O’Toole J C (2002) Yield responses of rice (Oryza sativa L.) genotypes to drought under rainfed lowlands: 2. Selection of drought resistant genotypes. Field Crops Res 73(2-3): 169-180.
- Ashley J (1993) Drought and crop adaptation. In: Rowland JRJ, editor, Dryland farming in Africa, Macmillan Press Ltd, UK, p. 46–67.
- Turner NC (1979) Drought resistance and adaptations to water deficits in crop plants. Mussel H, Staples RC (Eds) Stress physiology of crop plants. Wiley Interscience, New York, USA, pp. 343-372.
- Babu RC (2010) Breeding for drought resistance in rice: an integrated view from physiology to genomics. Electron J Plant Breed 1:1133-1141.
- Fukai S, Cooper M (1995) Development of drought-resistant cultivars using physio morphological traits in rice. Field Crop Res 40(2): 67-86.
- Price AH, Cairns JE, Horton P, Jones HG, Griffiths H (2002) Linking drought-resistance mechanisms to drought avoidance in upland rice using a QTL approach: progress and new opportunities to integrate stomatal and mesophyll responses. J Exp Bot 53(371): 989-1004.
- Swamy BPM, Kumar A (2011) Sustainable rice yield in water short drought prone environments: conventional and molecular approaches. Lee Teang Shui (Ed) Irrigation systems and practices in challenging environments. Intech Press, Croatia, Philippines, USA, pp. 149-168.
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296(5565): 92-100.
- Yu J, Hu S, Wang J, Wong G, Li S, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. Ssp indica). Science 296(5565): 79-92.
- Ekanayake IJ, O’Toole JC, Garrity DP, Masajo TM (1985) Inheritance of root growth characters and their relations to drought resistance in rice. Crop Sci 25(6): 927-933
- Ashfaq M, Haider MS, Khan AS, Allah SU (2012) Breeding potential of the basmati rice germplasm under water stress condition. Afr J Biotechnol 11(12): 6647-6657.
- Panwar LL (2005) Line × tester analysis of combining ability in rice (Oryza sativa L.). Ind J Genet Plant Breed 65: 51-52.
- Rita B, Moti Ramani NK (2005) Study on gene action and combining ability in rice. Oryza 42: 153-155.
- Singh BN, Mackill DJ (1990) Genetics of leaf rolling under drought stress. In: Brar DS, editor. Proceedings of the 2nd International Rice Genetics Symposium. International Rice Research Institute, Manila, Philippines, USA, pp.159–66.
- Gnanasekaran M, Vivekanandan P, Muthuramu S (2006) Combining ability and heterosis for yield and grain quality in two-line rice (Oryza sativa L.) hybrids. Ind J Genet Plant Breed 66: 6-9.
- Michael G, Rangasamy P, Nadarajan N (2003) Assessing the best combinations in rice (Oryza sativa L.) suitable for drought prone areas of Tamil Nadu. Res Crop 4: 79-84.
- McCouch SR, Teytelman L, Xu Y (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativaL.). DNA Research 9(6): 199-207.
- Temnykh S, Declerck G, Lukashova A, Lipovich L, McCouch, et al. (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations and genetic marker potential. Genome Res 11(8): 1441-1452.
- Tuberosa R (2004) Molecular approaches to unravel the genetic basis of water use efficiency. Bacon MA (Ed) Water use efficiency in plant biology. Blackwell, Oxford, UK, pp. 228-301.
- Tuberosa R, Salvi S (2006) Genomics-based approaches to improve drought tolerance of crops. Trends Plant Sci 11(8): 405-412.
- Tsonev S, Todorovska E, Avramova V, Kolev S, Abhu-Madhi N, et al. (2009) Genomics assisted improvement of drought tolerance in maize: QTL approaches. Biotechnology 23: 1410-1413.
- Xing YZ, Tan YF, Hua JP, Sun XL, Xu G, et al. (2002) Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet 105(2- 3): 248-257
- Collard BCY, Vera Cruz CM, McNally KL, Virk PS, Mackill DJ (2008) Rice molecular breeding laboratories in the genomics era: current status and future considerations. International Journal of Plant Genomics p. 25.
- Sasaki T, Burr B (2000) International rice genome sequencing project: the effort to completely sequence the rice genome. Curr Opin Plant Biol 3(2): 138-142.
- Tao Q, Chang YL, Wang J, Chen H, Islam Faridi MN, et al. (2001) Bacterial artificial chromosome-based physical map of the rice genome constructed by restriction fingerprint analysis. Genetics 158(4): 1711- 1724.
- McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, et al. (2009) Genome-wide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci U S A 106(30): 12273-12278.
- McCouch SR, Zhao K, Wright M, Tung CW, Ebana K, et al. (2010) Development of genome-wide SNP assays for rice. Breed Sci 60(5): 524- 535.
- Salvi S, Tuberosa R (2005) To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci 10(6): 297-304.
- Suji KK, Joel AJ (2010) An epigenetic change in rice cultivars under water stress conditions. Electron J Plant Breed 1(4): 1142-1143.
- WS, Pan YJ, Zhao XQ (2011) Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L) J Exp Bot 62(6): 1951-60.
- Langridge P, Paltridge N, Fincher G (2006) Functional genomics of abiotic stress tolerance in cereals. Brief Funct Genomic Proteomic 4(4): 343-354.
- Shimamoto K, Kyozuka J (2002) Rice as a model for comparative genomics of plants. Annu Rev Plant Biol 53: 399-419.
- The Rice Full-Length cDNA Consortium. (2003) Collection, Mapping, and Annotation of over 28,000 cDNA clones from japonica rice. Science 301(5631): 376-379.
- Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, et al. (2008) Genomewide analysis for discovery of rice microRNAs reveals natural antisense micro RNAs (nat-miRNAs). Proc Natl Acad Sci U S A 105(12): 4951-4956.
- Shen J, Xie K, Xiong L (2010) Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of micro RNAs in abiotic stress responses. Mol Genet Genomics 284(6): 477-488.
- Fu BY, Xiong JH, Zhu LH, Zhao XQ, Xu HX, et al. (2007) Identification of functional candidate genes for drought tolerance in rice. Mol Genet Genomics 278(6): 599-609.
- Bajaj S, Mohanty A (2005) Recent advances in rice biotechnology-- towards genetically superior transgenic rice. Plant Biotechnol J 3(3): 275-307.
- Stewart CN, Richards HA, Halfhill MD (2000) Transgenic plants and biosafety: science, misconceptions and public perceptions. Biotechniques 29(4): 832-836, 838-843.
- Tyagi AK, Mohanty A (2000) Rice transformation for crop improvement and functional genomics. Plant Sci 158(1-2): 1-18.
- Mochida K, Shinozaki K (2011) Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol 52(12): 2017-2038.
- Takeda S, Matsuoka M (2008) Genetic approaches to crop improvement: responding to environmental and population changes. Nat Rev Genet 9(6): 444-457.
- Jiang Y, Cai Z, Xie W, Long T, Yu H et al. (2012) Rice functional genomics research: progress and implications for crop genetic improvement. Biotechnol 30(5): 1059-1070.
- Yang W, Duan L, Chen G, Xiong L, Liu Q (2013) Plant phenomics and highthroughput phenotyping: accelerating rice functional genomics using multidisciplinary technologies. Curr Opin Plant Biol 16(2): 180-187.
- Rabello AR, Guimarães CM, Rangel PHN (2008) Identification of droughtresponsive genes in roots of upland rice (Oryza sativa L). BMC Genomics 9: 485.
- Skriver K, Mundy J (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2(6): 503-512.
- Kathiresan A, Lafitte HR, Chen J, Mansueto L, Bruskiewich R, et al. (2006) Gene expression microarrays and their application in drought stress research. Field Crop Res 97(1): 101-110.
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, et al. (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755-1767.
- Gorantla M, Babu PR, Reddy VB, Feltus FA, Patterson AH, et al. (2005) Functional genomics of drought stress response in rice: transcript mapping of annotated unigenes of indica rice (Oryza sativaL. cv. Nagina 22). Curr Sci 89(3): 496-514.
- Lenka SK, Katiyar A, Chinnusamy V, Bansal KC (2011) Comparative analysis of drought-responsive transcriptome in indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J 9(3): 315-327.
- Gao FH, Zhang HL, Wang HG, Gao H, Chao Z (2009) Comparative transcriptional profiling under drought stress between upland and lowland rice (Oryza sativa L.) using cDNA-AFLP. Chin Sci Bull 54: 3555- 3571.
- Jiao PY, Wang D, Zhu LH, Fu BY, Li ZK (2009) Differential expressions of two-component element genes in rice under drought stress. Acta Agron Sin 35(9): 1628-1636.
- Coudert Y, Périn C, Courtois B, Khong NG, Gantet P (2010) Genetic control of root development in rice, the model cereal. Trends Plant Sci 15(4): 219-226.
- Rebouillat J, Dievart A, Verdeil JL, Escoute J, Giese G, et al. (2009) Molecular genetics of rice root development. Rice 2(1): 15-34.
- Yang L, Zheng B, Mao C, Qi X, Liu F, et al. (2004) Analysis of transcripts that are differentially expressed in three sectors of the rice root system under water deficit. Mol Genet Genomics 272(4): 433-442.
- Wang H, Zhang H, Gao F, Li J, Li Z (2007) Comparison of gene expression between upland and lowland rice cultivars under water stress using cDNA microarray. Theor Appl Genet 115(8): 1109-1126.
- Chinnusamy V, Zhu JK (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12(2): 133-139.
- Makarevich G, Villar CB, Erilova A, Kohler C (2008) Mechanism of PHERES1imprinting in Arabidopsis. J Cell Sci 121(Pt 6): 906-912.
- Shibuya K, Fukushima S, Takatsuki H (2009) RNA-directed DNA methylation induces transcriptional activation in plants. Proc Natl Acad Sci U S A 106(5): 1660-1665.
- Zhao B, Liang R, Ge L, Li W, Xiao H, et al. (2007) Identification of droughtinduced microRNAs in rice. Biochem Biophys Res Commun 354(2): 585- 590.
- Soren K, Ali K, Tyagi V, Tyagi A (2010) Recent advances in molecular breeding of drought tolerance in rice (Oryza sativa L.). Indian J Biotechnol 9(3): 233-251.
- Krasensky J, Jonak C (2012) Drought, salt, and temperature stressinduced metabolic re-arrangements and regulatory networks. J Exp Bot 63(4): 1593-1608.
- Serraj R, Sinclair TR (2002) Osmolyte accumulation: can it really help increase crop yield under drought conditions? Plant Cell Environ 25(2): 333-341.
- Wang WS, Pan YJ, Zhao XQ (2011) Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L) J Exp Bot 62(6): 1951-60.
- Ji K, Wang Y, Sunb W, Lou Q, Mei H, et al. (2012) Drought-responsive mechanisms in rice genotypes with contrasting drought tolerance during reproductive stage. J Plant Physiol 169(4): 336-344.
- Mir RR, Zaman Allah M, Sreenivasulu N, Trethowan R, Varshney RK (2012) Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor Appl Genet 125(4): 625- 645.
- Vikram P, Swamy BPM, Dixit S, Ahmed HU, Sta Cruz MT, et al. (2011) qDTY1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet 18(12): 89.
- Zhao K, Tung CW, Eizenga GC, Wright MH, Ali LM, et al. (2011) Genomewide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2(13): 467.
- Xiong JH, Fu BY, Xu HX, Li YS (2010) Proteomic analysis of PEG-simulated drought stress responsive proteins of rice leaves using a pyramiding rice line at the seedling stage. Bot Stud 2(9): 137-145.
- Wang JH, Geng LH, Zhang C M (2012) Research on the weak signal detecting technique for crop water stress based on wavelet denoising. Adv Mat Res 424/425: 966-970.
- Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) A proteomic approach to analyzing drought- and salt-responsiveness in rice. Field Crop Res 76(2-3): 199-219.
- Rajasundaram D, Selbig J (2016) More effort--more results: recent advances in integrative “omics” data analysis. Current Opinion in Plant Biology 30: 57-61.
- Maksup S, Roytraku S, Supaibulwatana K (2012) Proteome and transcriptional responses in three contrasting indica rice (Oryza sativaL. ssp. indica) under water stress. Abstract of the 2012 CU-MUSC Graduate Forum in Plant Biotechnology.
- Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, et al. (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci USA 99(18): 11969-11974.
- Komatsu S, Tanaka N (2005) Rice proteome analysis: a step toward functional analysis of the rice genome. Proteomics 5(4): 938-949.
- Jorrín JV, Maldonado AM, Castillejo MA (2007) Plant proteome analysis: a 2006 update. Proteomics 7(16): 2947-2962.
- Choudhary M, Basu D, Datta A, Chakraborty N, Chakraborty S (2009) Dehydration-responsive nuclear proteome of rice (Oryza sativa L.) illustrates protein network, novel regulators of cellular adaptation, and evolutionary perspective. Mol Cell Proteomics 8(7):1579-1598.
- Ali GM, Komatsu S (2006) Proteomic analysis of rice leaf sheath during drought stress. J Proteome Res 5(2): 396-403.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.