Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Analysis of Somatic Variants in Growth Hormone Secreting Pituitary Adenomas by Whole Exome Sequencing
*Corresponding author: Young Zoon Kim, Division of Neurooncology and Department of Neurosurgery, Samsung Changwon Hospital, Sungkyunkwan University School of Medicine, 158, Paryong-ro, Masanhoewon-gu, Changwon, 51353, South Korea.
Received: August 22, 2019;Published: August 27, 2019
DOI: 10.34297/AJBSR.2019.04.000853
Abstract
Background: Whole-genome and whole-exome sequencing (WES) studies of pituitary adenoma have reported on average three mutations per patient tumor and discovered calcium signaling pathway mediated role of tumorigenesis. The aim of this study is to analyze the somatic variants in growth hormone (GH) secreting pituitary adenoma using WES.
Material and Methods: Whole-exome sequencing was used to identify somatic mutations in GH-secreting pituitary adenomas from 14 patients (7-45 years), including a sample from a patient with germline GPR101 duplication. A mean sequencing depth of 94x was achieved covering 98% of targeted exonic regions at a minimum depth of 10x.
Results: A total of 30 somatic variants were identified from 14 tumor samples. A recurrent hotspot mutation in GNAS, a well-established gene implicated in acromegaly, were identified in four patients. In addition, missense variants in CACNA1H and WIPI1 were identified which have only been reported in single patients previously. We also observed variants in GNAQ, RASGEF1B and PARP4 (this latter one in the GPR101-positive sample) genes that are involved in cAMP and calcium signaling pathways but have not been previously reported to be altered in acromegaly. Large deletions in chromosomes 1, 6, 10, 11, 12, 16, 17, 19 and 20 were also observed in at least eight patients.
Conclusion: By combining our findings with data from previous studies of GH-secreting adenomas, we conclude that pathways involving calcium and cAMP signaling are likely to be involved in the pathogenesis of somatotroph adenomas.
Keywords: Somatotropinoma; Acromegaly; Growth Hormone; Whole-Exome Sequencing
Introduction
Pituitary adenomas are usually benign, monoclonal neoplasms of anterior pituitary gland with an overall prevalence of 20% and clinically manifesting prevalence of 0.1% in the general population [1,2]. Invasive adenomas and carcinomas of the pituitary account for 35% and ~0.1-0.2% of all pituitary neoplasms, respectively, with the large majority being benign adenomas [2-4]. Although most pituitary adenomas are benign, the tumor size and hormonal status of the cell type that is affected ultimately determine the clinical phenotype. Prolactinomas are the most common (40-50%) hormone secreting pituitary tumors, followed by growth hormone (GH) secreting pituitary adenomas (15-20%). Other less common hormone secreting pituitary adenomas include adrenocorticotropin hormone (ACTH)-secreting adenomas, gonadotropin (lutein izing Hormone [LH] and follicle-stimulating hormone [FSH]) se creting adenomas, thyroid Stimulating Hormone (TSH)-secreting adenomas or mixed adenomas.
Clinically, pituitary tumors can present as sporadic or familial disease or syndromic or non-syndromic disease. The familial non-syndromic form (familial isolated pituitary adenomas (FIPA)) can be caused by AIP or GPR101 mutations, while familial syndromic forms are due to MEN1, PRKAC1A, SDHx, or rarely CDKN1B, PRKACB and DICER1 [5]. The majority (~93%) of pituitary tumors causing acromegaly are sporadic in nature with underlying genetic and epigenetic anomalies [6]. Two particular somatic gain-of-function mutations in GNAS, a gene encoding the α subunit of the stimulatory G protein, have been identified several decades ago, representing up to ~40% of somatotropinomas [7,8].
Identification of somatic genetic alternations in tumors is critical for understanding the molecular mechanism of tumorigenesis. The advancements in high-throughput sequencing and sophisticated analytical tools facilitate the unbiased genome-wide identification of somatic genetic mutations. Next-generation sequencing has been successful in elucidating the somatic mutational landscape of somatotropinomas [9-13], Cushing’s disease [14-16], nonfunctioning pituitary adenomas [17] and other subtypes of pituitary tumors [9-12].
Whole-genome and whole-exome sequencing (WES) studies of GH-secreting pituitary adenoma have reported on average three mutations per patient tumor and discovered calcium signaling pathway mediated role of tumorigenesis. The low rate of somatic mutations in pituitary tumors also indicates the possible role of other mechanisms of tumorigenesis involving differential miRNA expression [18], epigenetic alterations [19,20] and copy number variations [9,21,22]. Recurrent copy number aberrations including the whole-chromosomal losses are also commonly observed in pituitary adenoma [9,13,23].
In this study, we report somatic variant profiles of 14 somatotropinomas by WES. We identified a recurrent hotspot mutation in GNAS (4/14) and describe a deleterious somatic variant in CACNA1H, and likely benign variants in WIPI1, GNAS, GNAQ, RASGEF1B and PARP4.
Materials and Methods
Patient subjects and tissue samples
Patients diagnosed with GH-secreting pituitary adenomas at the Neurosurgical Department of Sungkyunkwan University Samsung Changwon hospital were recruited to the study. We analyzed the formalin-fixed, paraffin-embedded (FFPE) samples at the pathological archives which were obtained for recent 5 years. And the blood samples were routinely obtained before treatment including surgery and any medication at the department. All 14 patients provided written consent for use of their biological samples for diagnosis and research purposes. Pituitary tumors surgically resected at the time of endonasal endoscopic transsphenoidal surgery from the patients were collected, washed and frozen immediately. Peripheral blood from the patients was also collected and stored. Details of the patients including sex, age at diagnosis, pathological reports and levels of hormones for all patients are provided in Table 1. The median age of the patients at the time of diagnosis was 32 years (range 7-56).
DNA isolation and whole exome sequencing
FFPE tumor specimens and the peripheral blood collected from patients were subjected to genomic DNA isolation using DNeasy Blood and Tissue kit (QIAGEN, USA). The quality of genomic DNA was checked using the Agilent 2100 BioAnalyzer and the quantity was measured using NanoDrop and Qubit fluorometer 3.0. About 1 μg of genomic DNA was fragmented using Covaris LE220 for library preparation. The DNA fragment library was further enriched for targeted exome using probes against 357,999 exons spanning 21,522 genes in the human genome using SureSelectXT Human All Exon V5 capture kit (Agilent Technologies, USA). The quality of library was assessed using the Agilent 2100 BioAnalyzer. Indexed exome libraries from multiple samples were then pooled and subjected to paired-end sequencing with read length of 100 bp on an Illumina HiSeq 2500 platform. Raw data from the sequencing was then demultiplexed to generate fast q files for individual samples.
Whole exome sequencing data analysis to identifying somatic mutations
Sequencing data analysis was carried out using an in-house computational pipeline established following the guidelines previously established for variant discovery [24]. In summary, the sequencing quality of data was assessed using FastQC (v0.10.1) and all low-quality bases with Phred score <20 from read ends were soft masked from mapping against human genome primary assembly (GRCh37) using BWA (v0.7.10) [25]. The raw alignment was coordinate sorted using Samtools (0.1.19) [26], then processed to mask duplicate reads using MarkDuplicates (Picardtools v1.107) to filter for reads resulting from PCR duplicates during library preparation. Further steps of realignment around known indels and base quality score recalibration was carried out using Genome Analysis Toolkit (GATK v2.8-1) 27]. The somatic mutations were identified by comparing variants identified between pituitary tumor sample and corresponding blood samples for each patient using VarScan (v2.3.7) [28], and MuTect (v1.1.4) [29]. Sequence variants specifically observed in tumor DNA and completely absent in germline DNA were considered as somatic variants. Loss of heterozygosity in tumor genomes was detected using VarScan by comparing against the germline sequence. Annotation of identified somatic mutations was carried out using ANNOVAR (release: March 2015) [30] and SnpEff (v4.1) [31] against their curated reference databases for hg19. Variants with minor allele frequency (MAF) <0.01 were considered after comparing with variants from populations studies such as dbSNP, 1000 genomes project, EVS and ExAC [32]. Further, functional effects of the mutations were predicted using SIFT and Polyphen-2 scores. The shortlisted variants were validated by visualization of aligned data using integrated genomics viewer (IGV) [33].
Results and Discussion
Whole exome sequencing data analysis
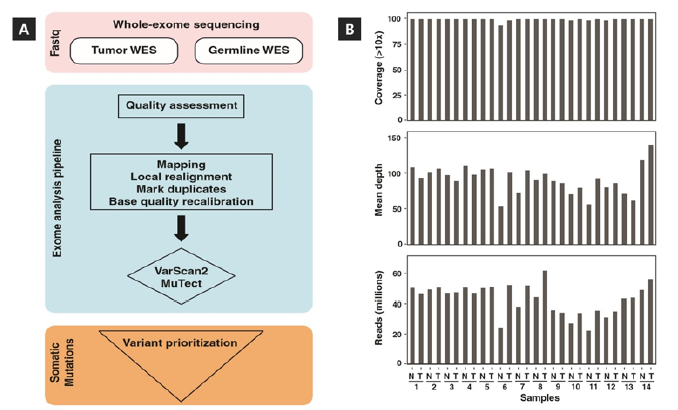
Figure 1: Schematic of data analysis workflow and alignment statistics. (A) Schematic workflow outlines the steps in whole-exome sequencing data analysis. The tumor and corresponding normal (germline) WES data were processed using an automated pipeline developed to implement the indicated workflow. Raw reads were aligned against human reference genome (hg19) and somatic variants were called to identify the mutations. (B) Plots shows the number of paired-end reads generated along with alignment depth and coverage for each patient sample.
A total of 14 tumor-blood sample pairs from acromegaly patients were selected for WES (Table 1). WES analysis of tumor and matched blood samples from patients were carried out to identify somatic mutations in GH-secreting pituitary adenomas. Approximately 43 million paired-end reads of length 100 bp were generated per sample. Quality analysis of data revealed >95% of the sequenced bases were with Phred quality score of ≥20 which is good for reliable detection of variants. Raw data generated from sequencing of samples were analyzed by using an in-house exome analysis pipeline to map against the human genome reference (hg19). A schematic representation of data analysis steps and tools used for the processing of raw and aligned data are depicted in Figure 1A. This analysis resulted in achieving a mean depth of 90x and average coverage of 98% over the targeted regions at minimum depth of 10x. A summary of sample sequencing data along with number of reads, depth and coverage is shown in Figure 1B. Overall, the data quality was sufficient for discovery of somatic variants in the tumors by comparing against the germline DNA of individual patients.
Somatic mutation profile of acromegaly
Next-generation sequencing of tumor genomes enables detection of driver genes that alter the physiology of cells to turn them cancerous. To identify somatic mutations in acromegaly, variants identified from tumor and normal patient genomes were compared. From the variant analysis of the 14 patient samples, we identified a total of 30 genes with somatic variants that alter their protein sequences and has a MAF<0.01. We observed up to 4 somatic changes per tumor across the 14 patient samples (Table 2). The most recurrent mutations were observed in the GNAS gene (5 out of 14 patients), which is a well-established gene known to be mutated in acromegaly. Four out of five patients carried the well-known hot spot mutation p.R201C in GNAS gene, while the fifth patient had a novel and predicted benign GNAS variant p.G553D. In addition, we identified somatic variants in CACNA1H, WIPI1, RASGEF1B, PARP4 and GNAQ genes (Table 2).
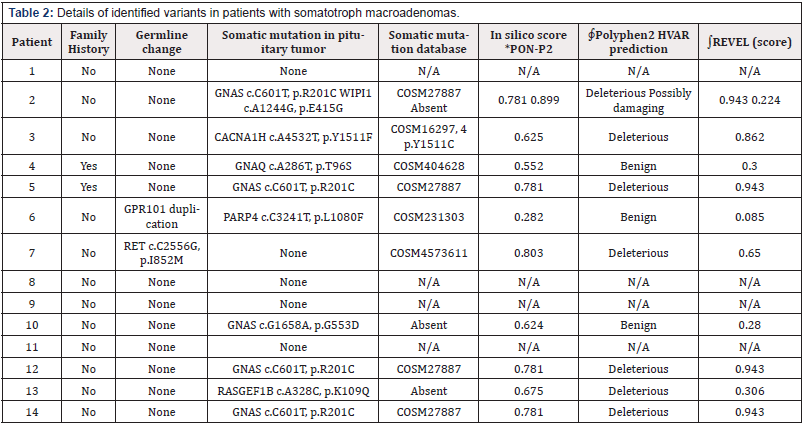
Table 2: Details of identified variants in patients with somatotroph macroadenomas.
*PON-P2 score is a probability score for pathogenicity of variant.
∮Polyphen2 predictions represent the possible impact of an amino acid substitution on the structure and function of a human protein.
∫REVEL score is based on an ensemble method for predicting the pathogenicity of missense variants on the basis of multiple individual prediction tools, the closer score to 1, the higher the chance the variant being damaging. Threshold usually taken at 0.75.
Interestingly, the CACNA1H p.Y1511F change in the calcium voltage-gated channel subunit alpha1 H has not been previously described and is rendered deleterious by several prediction programs. A different change affecting the same amino acid was seen in the COSMIC database (p.Y1511C). We found a novel missense change in WIPI1, also a calcium pathway protein playing a role in calcium- mediated autophagy. The RASGEF1B p.K109Q variant is rare and predicted to be deleterious by Polyphen, but not by REVEL. The RASGEF1B gene is a guanine-nucleotide exchange factor and known interactor of Ras family of proteins which are essential regulators of cell proliferation, survival, differentiation, actin organization, vesicular trafficking, and gene expression. The PARP4 p.L1080F somatic change was previously seen in the COSMIC database and predicted to be benign. Genetic changes seen in this study are listed in Table 2 and depicted in Figure 2A
Although WES has its limitations to detect large chromosomal variations, we compared tumor normal DNA sequencing data to identify large deletions or amplifications of chromosomal segments which were previously implicated in somatotroph tumorigenesis. Analysis of genome-wide loss of heterozygosity events in our WES data revealed somatic deletions in chromosomes of at least eight patients. Complete hemizygous deletion of chromosome 6 and 19 were observed in patient #3 and #12, respectively. Other small focal deletions in chromosome 1, 6, 10, 11, 12, 16, 17, 19 and 20 were observed in patients #2, #5, #9, #10, #13 and #14 as showed in Figure 2B. Previous studies of GH-secreting pituitary adenomas using next generation sequencing approach have also reported chromosome losses (chromosomes 1, 6, 13, 14, 15, 16, 18 and 22) and complex rearrangements [9-13]. In a recent unpublished study, increased somatic copy-number variation was associated with DNA damage resulting from enhanced cAMP activity in sporadic somatotroph adenomas [34].
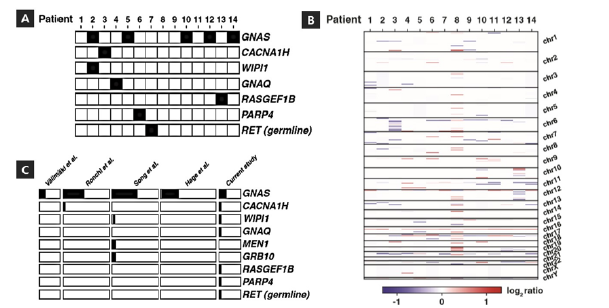
Figure 2: SSomatic changes observed in somatotrophs from WES analysis. (A) Deleterious mutations observed in individual tumors from patients are indicated as filled black squares along with the gene symbols of the corresponding genes. (B) Large structural changes observed from WES analysis across samples. (C) Mutated genes across GH-secreting pituitary tumors studied by various groups are indicated along with their relative frequencies represented as horizontal bars.
Patient #4 with a benign somatic GNAQ variant (not seen in germline but seen in COSMIC previously) is a FIPA case as her mother was also affected by acromegaly. No tissue sample is available from the mother. Patient #6 was positive for GPR101 duplication and the benign PARP4 variant was observed in the tumor. Patient #7 with a recurrent tumor was identified with a germline variant p.I852M in the RET gene. He has normal calcitonin, calcium and parathormone levels, and normal urinary metanephrins. This RET variant has been described in medullary thyroid carcinoma [35] and Multiple Endocrine Neoplasia 2A patients [36] and most likely represents a benign variant, despite the prediction scores [37]. Acromegaly has been described in a few patients with RET mutations, but these probably represent coincidences, rather than linked pathogenetically to the RET mutations [38,39]. Physical reexamination of a subset of patients with GNAS mutations in tumor did not reveal any skin lesions and their bone scans were normal, therefore ruling out McCune-Albright syndrome. The presence of variants was not correlating to the sparsely or densely granulated nature of the tumours, or to size and invasiveness.
Mutation data in five independent cohorts provided an opportunity to compare our results to the genetic profiles of other somatotropinomas (Figure 2C). A summary and a partial list of genes with variants reported by these recent publications are provided in Table 2 & 3. Mutations in GNAS are the only mutations recurrent (Figure 3A). The identification of variants in CACNA1H and WIPI1 in this study and a previous study, together with other calcium pathway protein changes emphasize the key role of calcium signaling in tumorigenesis of GH-secreting pituitary adenomas (Figure 3B & 3C). Sequencing of additional pituitary tumors in the future may lead to discovery of new driver genes contributing to understanding of tumorigenesis in acromegaly.
Global comparison of variants across various pituitary adenomas
Clinically non-functioning pituitary adenomas were analyzed in three studies [9,12,17] which did not identify any recurrent mutations. While recurrent GNAS mutations at two sites (codon 201 and 227) are seen in somatotroph adenomas, and recurrent USP8 mutations at the binding site of 14-3-3 protein in corticotroph adenomas, truncating mutations of NR3C1 was observed in adrenocorticotroph adenomas and MEN1 somatic loss-of-function changes in plurihormonal adenomas producing growth hormone and prolactin [12]. The majority of the identified mutations in these genes were observed in known hot spot regions and only a few were novel. To enable general observations in somatotroph tumors, we compared the mutation data from current study to all five recent studies. Mutations in CACNA1H and WIPI1 were seen in only two patients with GH-hypersecretion including cases from this study. Non-overlapping genetic mutation profiles between various subtypes of pituitary adenomas suggests a possible role of distinct mechanisms of tumorigenesis and secretion of hormones. This comparison suggests a low mutation burden across the pituitary tumors and mutations in classical cancer genes such as TP53 and RAS are almost never seen [5].
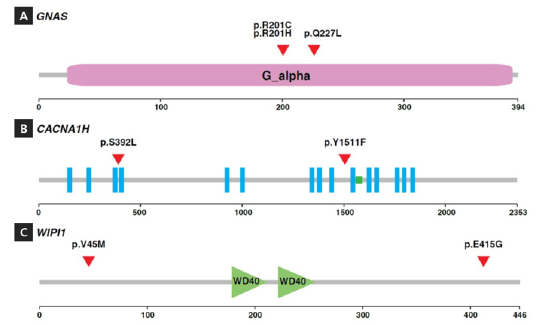
Figure 3: Mutations identified in genes with multiple mutations. The domain structures along with the locations of the substitutions (marked by red triangles) are shown for GNAS (A), CACNA1H (B) and WIPI1 (C) The scale at the bottom marks’ amino acid positions in the protein sequences.
Biological relevance of genes with variants
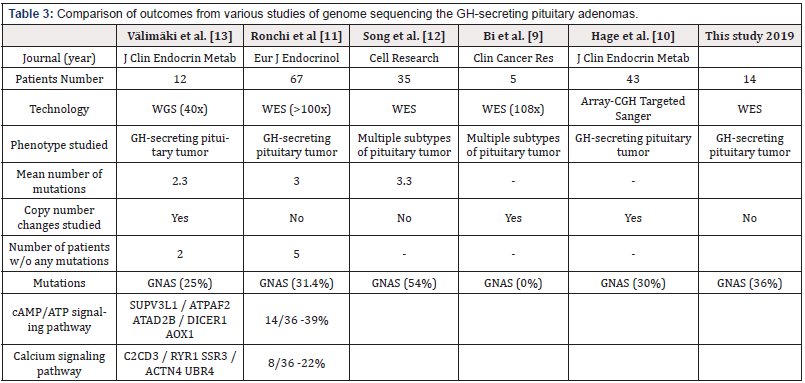
Table 3: Comparison of outcomes from various studies of genome sequencing the GH-secreting pituitary adenomas.
CGH: Comparative Genomic Hybridization; GH: Growth Hormone; WES: Whole Exome Sequencing; WGS: Whole Genome Sequencing
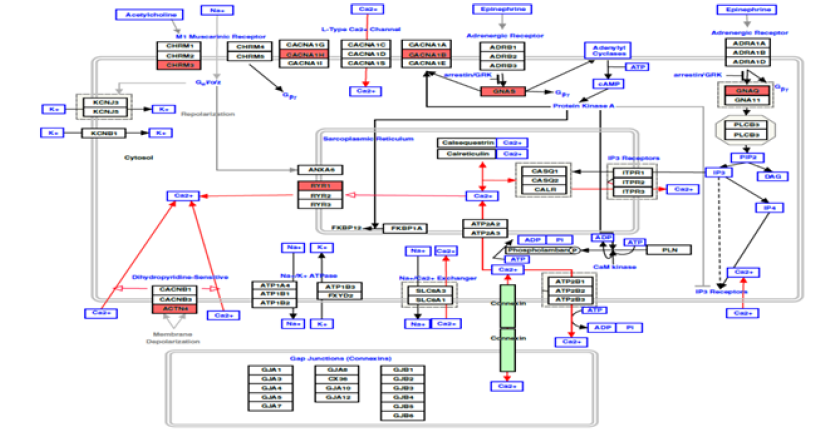
Activated calcium and cAMP signaling pathways lead to an increase in cytosolic free calcium which triggers secretion of GH. In addition, ATP released along with pituitary hormones induces intracellular free calcium accumulation which in turn leads to secretion of GH [40]. Recurrent mutations of GNAS, CACNA1H and WIPI1 genes suggest a vital role of cAMP and calcium signaling in acromegaly. Activating mutations identified in GNAS gene were associated with production of cAMP from ATP by activation of adenylate cyclase. CACNA1H is a member of alpha-1 subunit family of the voltage-dependent calcium channel complex and directly mediates calcium flux. WIPI1 plays an important role in calcium-mediated autophagy during increased levels of cytosolic calcium. The Ras protein family interactor RASGEF1B is a novel gene found with changes in this tumor type and mutations of PARP4 were previously reported in thyroid and breast cancers. Overall, a total of 15 and 12 genes involved in cAMP and calcium signaling, respectively were found changed in somatotropinomas as observed in current or previous studies (Table 3) & 4). A pathway diagram of calcium signaling adopted from Wiki-Pathways (https://www.wikipathways.org/ instance/WP536) is presented in Figure 4 with the mutated genes highlighted in red. Identification of mutations across several of these genes strongly suggest the dysregulation of the calcium and cAMP signaling which are potentially involved in pituitary tumorigenesis (Table 1).
Conclusion
Somatotroph adenomas are mostly driven by somatic mutations. Next-generation sequencing-based platforms enable largescale analysis of patient samples to decipher mutational landscapes of tumors. We undertook WES of GH-secreting pituitary tumors, including one with germline GPR101 duplication, to detect somatic changes. We identified recurrent mutations in GNAS, a well-established gene implicated in causing acromegaly, in four out of 14 patients. Combined analysis of somatic variants from this study with recent studies on somatotropinomas identified two additional candidate genes of importance, CACNA1H and WIPI1 in two independent cohorts. Variants in other associated genes GNAQ, RASGEF1B and PARP4 were also identified in individual samples. No somatic mutations of current relevance were identified in five tumors, although large deletions were present in two of them. Our data suggest that whole-exome sequencing have limitations identifying definite cause of pituitary adenomas and points to the direction of studying epigenetic changes, structural changes or whole genome sequencing should to fully understand somatotroph tumorigenesis.
Disclosure/Conflict of Interest
The authors report no competing financial interests or conflicts of interest.
References
- Ezzat S, Asa SL, Couldwell WT, Barr CE, Dodge WE, et al. (2004) The prevalence of pituitary adenomas: a systematic review. Cancer 101(3): 613-619.
- Fernandez A, Karavitaki N, Wass JAH (2010) Prevalence of pituitary adenomas: a community-based, cross-sectional study in Banbury (Oxfordshire, UK). Clin Endocrinol (Oxf) 72(3): 377-382.
- Herman V, Fagin J, Gonsky R, Kovacs K, Melmed S (1990) Clonal Origin of Pituitary Adenomas. J Clin Endocrinol Metab 71(6): 1427-1433.
- Scheithauer BW, Kovacs KT, Laws ER, Randall RV (1986) Pathology of invasive pituitary tumors with special reference to functional classification. J Neurosurg 65(6): 733-744.
- Caimari F, Korbonits M (2016) Novel Genetic Causes of Pituitary Adenomas. Cancer Res 22(20): 5030-5042.
- Aflorei ED, Korbonits M (2014) Epidemiology and etiopathogenesis of pituitary adenomas. J Neurooncol 117(3): 379-394.
- Spada A, Arosio M, Bassetti M, Vallar L, Clementi E, et al (1991) Mutations in the alpha subunit of the stimulatory regulatory protein of adenylyl cyclase (Gs) in human GH-secreting pituitary adenomas. Biochemical, clinical, and morphological aspects. Pathol Res Pract 187(5): 567-570.
- Vallar L, Spada A, Giannattasio G (1987) Altered Gs and adenylate cyclase activity in human GH-secreting pituitary adenomas. Nature 330(6148): 566-568.
- Bi WL, Horowitz P, Greenwald NF, Abedalthagafi M, Agarwalla PK, et al. (2017) Landscape of Genomic Alterations in Pituitary Adenomas. Clin Cancer Res 23(7): 1841–1851.
- Hage M, Viengchareun S, Brunet E, Villa C, Pineau D, et al. (2018) Genomic Alterations and Complex Subclonal Architecture in Sporadic GH-Secreting Pituitary Adenomas. J Clin Endocrinol Metab 103(5): 1929-1939.
- Ronchi CL, Peverelli E, Herterich S, Weigand I, Mantovani G, et al (2016) Landscape of somatic mutations in sporadic GH-secreting pituitary adenomas. Eur J Endocrinol 174(3): 363-372.
- Song ZJ, Reitman ZJ, Ma ZY, Chen JH, Zhang QL, et al. (2016) The genome- wide mutational landscape of pituitary adenomas. Cell Res 26(11): 1255-1259.
- Välimäki N, Demir H, Pitkänen E, Kaasinen E, Karppinen A, et al. (2015) Whole-Genome Sequencing of Growth Hormone (GH)-Secreting Pituitary Adenomas. J Clin Endocrinol Metab 100(10): 3918-3927.
- Cao Y, He M, Gao Z, Peng Y, Li Yanli, et al. (2014) Activating hotspot L205R mutation in PRKACA and adrenal Cushing’s syndrome. Science 344(6186): 913-917.
- Perez-Rivas LG, Theodoropoulou M, Ferraù F, Nusser C, Kawaguchi K, et al. (2015) The Gene of the Ubiquitin-Specific Protease 8 Is Frequently Mutated in Adenomas Causing Cushing’s Disease. J Clin Endocrinol Metab 100(7): E997-E1004.
- Reincke M, Sbiera S, Hayakawa A, Theodoropoulou M, Osswald A, et al. (2015) Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat Genet 47(1): 31-38.
- Newey PJ, Nesbit MA, Rimmer AJ, Head RA, Gorvin CM, et al (2013) Whole-Exome Sequencing Studies of Nonfunctioning Pituitary Adenomas. J Clin Endocrinol Metab 98(4): E796-E800.
- Bottoni A, Zatelli MC, Ferracin M, Tagliati F, Piccin D, et al. (2007) Identification of differentially expressed microRNAs by microarray: A possible role for microRNA genes in pituitary adenomas. J Cell Physiol 210(2): 370-377.
- Kober P, Boresowicz J, Rusetska N, Maksymowicz M, Goryca K, et al. (2018) DNA methylation profiling in nonfunctioning pituitary adenomas. Mol Cell Endocrinol 473: 194-204.
- Zhou Y, Zhang X, Klibanski A (2014) Genetic and epigenetic mutations of tumor suppressive genes in sporadic pituitary adenoma. Mol Cell Endocrinol 386(1-2): 16-33.
- Melmed S (2011) Pathogenesis of pituitary tumors. Nat Rev Endocrinol 7(5): 257-266.
- Szymas J, Schluens K, Liebert W, Petersen I (2002) Genomic Instability in Pituitary Adenomas. Pituitary 5(4): 211-219.
- Pack SD, Qin LX, Pak E, Wang Y, Ault DO, et al. (2005) Common genetic changes in hereditary and sporadic pituitary adenomas detected by comparative genomic hybridization. Genes Chromosomes Cancer 43(1): 72-82.
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics 43: 491-498.
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows- Wheeler transform. Bioinforma Oxf Engl 25(14): 1754-1760.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAM tools. Bioinforma Oxf Engl 25(16): 2078-2079.
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9): 1297-1303.
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, et al. (2012) Var- Scan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22(3): 568-576.
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, et al. (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31(3): 213-219.
- Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16): e164-e164.
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2): 80-92.
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616): 285-291.
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29(1): 24-26.
- Ben Shlomo A (2019) Functional Genomic Profile of Sporadic Pituitary Adenomas, in: Pituitary Congress Programs & Abstracts, Translational Science. Presented at the Sixteenth International Pituitary Congress, New Orleans, Louisiana, USA. p. 15.
- Demeester R, Parma J, Cochaux P, Vassart G, Abramowicz MJ (2001) A rare variant, I852M, of the RET proto-oncogene in a patient with medullary thyroid carcinoma at age 20 years. Hum Mutat 17(4): 354.
- Mathiesen JS, Kroustrup JP, Vestergaard P, Stochholm K, Poulsen PL, et al. (2016) Distribution of RET Mutations in Multiple Endocrine Neoplasia 2 in Denmark 1994–2014: A Nationwide Study. Thyroid 27(2): 215-223.
- Joshi RR, Heineman TE, Kutler DI, Cohen MA, Kuhel WI (2016) Multiple endocrine neoplasia type 2 kindred with novel tandem RET mutations: Case report with an applied in silico mutational tolerance analysis. Head Neck 38(Suppl 1): E1881-E1885.
- Brauer VF, Scholz GH, Neumann S, Lohmann T, Paschke R, et al. (2004) RET germline mutation in codon 791 in a family representing 3 generations from age 5 to age 70 years: should thyroidectomy be performed.Am Assoc Clin Endocrinol 10(1): 5-9.
- Saito T, Miura D, Taguchi M, Takeshita A, Miyakawa M, et al. (2010) Coincidence of multiple endocrine neoplasia type 2A with acromegaly. Am J Med Sci 340(4): 329-331.
- Canny BJ, Rawlings SR, Leong DA (1992) Pituitary adenylate cyclase-activating polypeptide specifically increases cytosolic calcium ion concentration in rat gonadotropes and somatotropes. Endocrinology 130(1): 211-215.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.