Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Detection of Resistance mecA Gene In Gram Positive Bacteria Described as Nosocomial
*Corresponding author: Navarro C, Department of Preventive Animal Medicine, University of Chile, Chile
Received: August 08, 2019 Published: August 21, 2019
DOI: 10.34297/AJBSR.2019.04.000831
Abstract
Shortly after the introduction of an antimicrobial to the market it has been possible to find bacteria that are resistant to its action. This resistance is natural in some bacteria, while in others it is a condition acquired through the incorporation of genes that code various mechanisms of resistance.
These resistant strains represent a big problem in human hospitals when causing nosocomial infections, since the therapeutic options are limited. In veterinary medicine, although nosocomial infections are increasing, they are still less studied than those acquired by people. However, these infections -both in human and veterinary patients- have in common to be caused, mainly, by methicillin-resistant staphylococci. Due to this fact, in addition to the fact that methicillin-resistant Staphylococcus aureus is transmitted between different animal species, including humans, the purpose of this work was to detect the mecA resistance gene in bacteria described as nosocomial.
For this, three environmental bacterial strains isolated from Veterinary Hospitals of the University of Chile and that showed resistance to oxacillin were used: Staphylococcus kloosii, Micrococcus sedentarius and Enterococcus faecium. After the extraction of the DNA, the technique of the Polymerase Chain Reaction and the corresponding electrophoresis, fluorescent bands of approximately 500 bp were obtained. These amplicons were sent to be sequenced and the sequences obtained were compared with those described in the GenBank® for the mecA gene. The percentages of nucleotide identity of 97%, 97% and 98%, respectively, confirm the presence of strains that possess the mecA gene and suggest its early incorporation as positive control strains in future investigations.
Keywords: Bacterial resistance; Nosocomial infection; mecA gene
Background
Nosocomial infections correspond to those infections that are acquired within a hospital ward and whose manifestation, depending on the incubation period of the infection, can occur 48 or 72 hours later, or even once the patient is discharged [1,2]. For them to occur, the characteristics of the microorganism in question must be considered, as well as those of the patient and the environment in which he or she is found. In addition, if it occurs, it is most likely that the microorganism has been acquired from poorly disinfected/sterilized material or from the personnel itself, which acts as a reservoir for many bacterial strains, being one of them, and the main cause of nosocomial infections described, methicillin-resistant Staphylococcus aureus (MRSA) [3,4]. The clinical isolates obtained from these patients are often resistant to antibiotics, especially those most frequently used in that hospital. Moreover, many times these species are shown as multi-resistant. In any case, whether infections caused by bacteria resistant to one type of antimicrobial or several of them, the result will be the same: increase in morbidity, mortality and hospital costs [5,6].
A bacterium resistant to antibiotics is considered when it can survive facing therapeutic concentrations of this drug. This resistance becomes especially important when it is present in pathogenic bacterial genera, since the treatment of the infections that they cause will be difficult. From the mechanisms of bacterial resistance described - enzymatic inactivation of the antibiotic, modification of the permeability of the bacterial membrane, alteration or production of new target sites and presence of efflux pumps - the bacteria can express one, more than one or all of them in a manner simultaneous [7,8].
In addition, this characteristic of being resistant to one or more antimicrobials is, for some bacteria, something intrinsic. This does not necessarily imply that the others are always sensitive, because in addition to chromosomal mutations, there is the possibility that a bacterium acquires an extrachromosomal DNA fragment - plasmids, transposons, integrons and / or genetic cassettes - carrier of genes that modify the resistance to the antibiotic, through the processes of transduction, transformation and / or conjugation [9-11].
In 1928 Alexander Fleming discovered the first natural antibiotic produced by the fungus Penicillium notatum, penicillin, which represented a breakthrough for the treatment of systemic infections caused by Staphylococcus aureus. However, given the limitations of this drug, others have been semisynthesized to achieve better results in the treatments. Hence, there are currently acid-fast penicillins, penicillinase-resistant penicillins, broad spectrum penicillins, penicillins active against Pseudomonas aeruginosa and penicillins with a long-acting profile [12,13]. The mechanism of action of β-lactams is the inhibition of the synthesis of peptidoglycan, constituent of the bacterial cell wall. This structure is formed by linear fibers of N-acetylmuramic acid and N-acetylglucosamine, which are cross-linked by the action of Penicillin-binding Proteins (PLP) to form a stable wall. These proteins bind to the penicillins, which have a structure similar to the terminal end of the pentapeptide linked to N-acetylmuramic acid (D-alanine-D-alanine), inactivating irreversibly, which weakens the cell wall, causing osmotic lysis of the bacteria [14,15].
The chemical structure of penicillin has a β-lactam ring associated with a thiazolidinic ring of 5 carbon atoms, thus forming 6-aminopenicillanic acid. A side chain is attached to the β-lactam ring -responsible for biological activity- which determines the antibacterial and pharmacokinetic characteristics, such as the antibacterial spectrum, resistance to penicillinase and the acid environment. It is this side chain that is modified to semisynthesize the other antimicrobials of this group [14,15]. Only the first three resistance mechanisms mentioned above are applicable to β-lactams, with enzymatic inactivation by β-lactamase production being the most important. For this reason, it was necessary to create antistaphylococcal penicillins, such as methicillin and oxacillin, that resist the action of β-lactamases. However, after the introduction of these drugs in clinical practice, methicillin-resistant staphylococci were found. In these bacteria, the target site of the β-lactams was modified [16,12,17].
PLP2 is normally expressed in the cell wall and has great affinity for methicillin. However, in strains isolated from methicillinresistant S. aureus (MRSA), a variant with very low affinity for this drug was found, but it is capable of carrying out all the required cellular functions. This PLP2a or PLP2 ‘is encoded by the mecA gene, which is part of a mobile complex (mec) and resides in a genetic element called Staphylococcal Chromosome Cassette in S. aureus (SCCmec). In this way, MRSA isolates are resistant to all β-lactam antibiotics and, frequently, to many other classes of antimicrobials [12,18].
Although in the beginning MRSA infections were confined to the hospital environment (SAMR-AH), today it is possible to find community-associated infections (MRSA-AC), which happened after being caused by patients after hospital discharge, the same health care personnel and visitors -who would have acted as vectors- to have an unclear origin [19,20].
So far, the difference that has been established between both types of MRSA lies in the spectrum of antimicrobials to which they are resistant. While SAMR-AC is resistant only to β-lactams, because it has the SCCmec type IV (of small size, so it can only harbor the mecA gene), SAMR-AH -which can have types I, II or III of SCCmechas been shown to be multiresistant, so much so that vancomycin is often the only therapeutic alternative. However, strains showing a reduced sensitivity to vancomycin have already been reported [18].
The origin of staphylococcal resistance to β-lactams in dogs and cats is not clear, since, unlike what happened in human medicine, methicillin and / or the other anti-staphylococcal penicillins have not been used in them. However, it must be considered that the mecA gene is in a mobile genetic element, so it can be transferred to other bacteria [21-23].
In this way, Staphylococcus pseudointermedius, which is part of the normal skin flora of dogs, but potentially pathogenic and zoonotic, has been shown to have the same resistance genes as S. aureus of human origin, suggesting a possible genetic exchange between the two bacterial species [24,25].
Although MRSA infections in companion animals are not frequent, since the first isolate of this bacterium was obtained in dogs in 1994, the number of these infections has increased, which has been attributed to the increase of SAMR-AC. In fact, the transmission of MRSA has been from the human being to companion animals, which can eliminate the microorganism, become infected or remain as a reservoir. In this way, both dogs and cats, healthy and sick, can be a new source of infection for other domestic animals and other people -like the veterinary staff that attends themincluding reinfection to their owners [24,18].
For all the above described, the general objective of this Title Memory was to detect the mecA resistance gene in Gram positive bacteria isolated from hospital enclosures and described as nosocomial, and the specific objectives to detect and sequence a fragment suspected to be constitutive of the mecA gene. in these bacteria, in addition to determining the percentage of nucleotide identity of the obtained fragment with respect to official GenBank® data.
Material and Methods
This research was carried out in the Laboratory of Veterinary Microbiology, belonging to the Department of Animal Preventive Medicine of the Faculty of Veterinary and Animal Sciences of the University of Chile. Samples Three bacterial strains phenotypically resistant to Oxacillin were studied, according to the Kirby- Bauer plate diffusion method, obtained in a previous study [26] Enterococcus faecium, Staphylococcus kloosii and Micrococcus sedentarius (Figure 1).
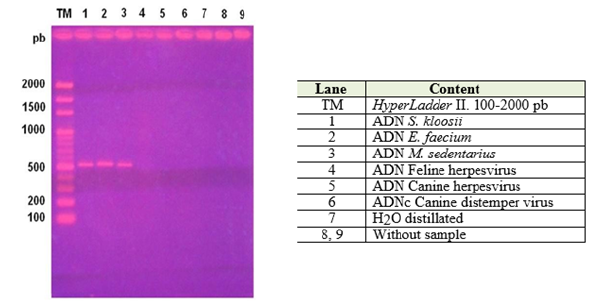
Figure 1: Electrophoresis in 2% agarose gel. Detection of gen mecA in 3 bacterial strains described as Nosocomials.
Detection of mecA gene by the Polymerase Chain Reaction (PCR)
Obtaining the bacterial DNA: The extraction of bacterial DNA was carried out by using a commercial kit (Genomic DNA Purification kit, Fermentas®). From cultures of 1.5x108 CFU/mL (obtained by turbidity compared to the 0.5 tube of McFarland) 200μl were taken, to which 400μl of lysis solution was added, being incubated for five minutes at 65° C. Then, they were added 600μl of chloroform, mixing gently and centrifuged at 10,000 rpm for two minutes (Heraeus Sepatech Biofuge®). Subsequently, the upper phase was collected in an Eppendorf tube and 800μL of precipitation solution was added, to be centrifuged again at 10,000 rpm for two minutes. The sediment obtained was resuspended with 100μl of 1.2 M sodium chloride solution and vortexed. To this mixture was added 300μl of cold ethanol, to be kept at -20° C for ten minutes. It was centrifuged again at 10,000 rpm for four minutes, then removing the supernatant and resuspending the pellet obtained in 100μl of nuclease-free water (Winkler®).
PCR: A kit (2X PCR Master Mix, Fermentas®) containing thermostable polymerase, deoxynucleotide triphosphates (dNTPs), reaction buffer and MgCl2 was used, from which 15μL was extracted to be deposited in a 0.2mL Eppendorf tube, together with 5μL of each of the primers (Annex, Table 1 & 2) and 5μL of the DNA sample, obtaining a total volume of 30μL.
DNA amplification: An Apollo thermocycler was used, with an initial denaturation at 94° C for one minute. Subsequently, 30 cycles of amplification with denaturation at 94º C for one minute, alignment at 57º C for one minute and elongation at 72º C for two minutes. Finally, a final elongation at 72º C for five minutes. With this methodology, a fragment of 533 bp [27] was obtained.
Visualization of the amplified products: It was performed by electrophoresis in 2% agarose gel (Winkler®) in Tris acetate EDTA buffer (TAE, Fermentas®). The PCR product was mixed with 1μl of the commercial loading product, 6X MassRuler DNA Loading Dye Solution (Fermentas®), which has glycerol -which gave density to the sample- and bromophenol blue to verify the migration of the bands of DNA An aliquot of 6μl of this mixture was deposited in the respective well of the gel. A standard that contains DNA fragments ranging between 100 and 2000 bp (HyperLadder II, Bioline®) was used as a molecular size marker. The electrophoresis was carried out at 90 V for 90 minutes and then the gel was incubated in ethidium bromide (0.5 μg /mL) (Fermelo®) for thirty minutes and then submerged in distilled water for ten minutes, after which the bands they could be visualized in a transilluminator of ultraviolet light (Transiluminator UVP®) and photographed.
Biosafety measures: The laboratory work was carried out according to the biosafety levels established for the Microbiology laboratory, such as the use of sterile material, closed apron and gloves, especially when handling the gel with ethidium bromide, in addition to the correct waste disposal. Since the visualization process of the amplified product involved the use of a UV light transilluminator, it was necessary to use an acrylic plate and glasses with UV filter. Once the photograph was obtained, the gel was incinerated.
Determination of nucleotide identity percentage (NIP): The DNA fragments were sent to the Sequencing Center of Genytec Ltda. To obtain the respective sequences. Subsequently, they were analyzed using the free access online ClustalW program, which aligned the obtained sequences and allowed obtaining a consensus sequence for each sample used, the latter being those that were compared with the mecA gene from GenBank accession number BX571856 through the same program, which allowed to establish, finally, the percentage of nucleotide identity.
Results
Detection and sequencing of fragments suspected to be constitutive of the gene mecA
The three analyzed samples presented a visible band in the UV light transilluminator, which were located around the 500 bp fragment of the molecular size marker (Figure 1), so that the amplicons obtained were sent to sequence. Determination of the NIP of each fragment obtained with respect to the official database (GenBank®). The nucleotide sequences delivered by Genytec Ltda. Were entered the open access online ClustalW program, which aligned them and allowed obtaining the consensus sequences (Tables 3-5), which, later, were compared with the sequence of the mecA gene that is in the official data bank, through the same program which gave them a NIP of between 97% and 98% .
Discussion
Humans and companion animals, given the close contact that exists between them, tend to transfer to each other bacteria, which may possess antimicrobial resistance genes. One of these genes is mecA, which, because it is part of a mobile genetic element, has spread among the different species of staphylococci [24]. Due to the heterogeneous nature of this gene expression, many bacterial strains may appear as sensitive to β-lactams by the Kirby-Bauer method, however, having the mecA gene, hence the detection of this and other resistance genes. by PCR it is very helpful [16].
When analyzing the results obtained, it was possible to verify that the processing of the three samples was successful. In this sense, the PCR protocol applied was adequate, since amplicons of approximately 500 bp were generated (Annex, Figure 1), which is a good indication of the presence of the gene and is consistent with a choice of optimal primers, so that it is not necessary to design new ones [27]. However, this is not sufficient to ensure the presence of a fragment or the totality of a particular gene. It should be mentioned that the primers only recognize a small complementary region and not all the fragment sought. That is, the nucleotide sequence found between the binding sites of the primers could correspond to a nonspecifically amplified sequence, so it is relevant to know the consensus nucleotide sequence of the 500 bp fragment. It should be noted that not unspecific bands of other molecular weights were observed when using the respective negative controls (DNA herpesvirus and cDNA distemper virus) or to use distilled water, which acts sentinel Accidental contamination of reagents PCR (Mastermix and primers)
Malik and colleagues in 2006, compared fragments of the mecA gene of methicillin-resistant staphylococci isolated from dogs and cats, both healthy and sick, obtaining identical nucleotide sequences. Subsequently, in [28], Bemis and colleagues also compared mecA genes, but only of S. pseudointermedius, which were found to be the same among them and with the sequences available in the GenBank® of S. haemolyticus and S. aureus, but different in 1 and 2 nucleotides to the sequences described in the same database for S. pseudointermedius. Although in this study a NIP of 100% was not achieved among the consensus sequences for each of the strains, they did not differ in more than one 3%, which suggested that it is the same gene in the three bacterial strains (Annex).
The nucleotide alignment of the consensus sequences versus a sequence stored in the GenBank® yielded high values of NIP in the three strains studied: 97% for both S. kloosii and M. sedentarius, and 98% for E. faecium, which confirms that this gene is highly conserved, as suggested [16]. Although this alignment was made using the Clustal W on-line program, there is an alternative analysis: the BLAST program (Basic Local Alignment Search Tool) that deals with the sequence obtained from a panel of sequences from an official database. When entering the sequences obtained in the BLAST program, they show a nucleotide identity of at least 97% with various variants of the mecA gene, whose sequences are stored in an extensive database (Annex). Thus, both programs endorse the NIP obtained.
The results reflect in a certain way the reality in veterinary medicine, in relation to strains participating in nosocomial infections and the resistance genes that they possess.
These results complement a previous project in the Microbiology Laboratory, in which several resistance genes were detected: blaTEM, in case of β-lactams; tetA and tetB, tetK, tetM and tetO in the case of tetracyclines; vanA and vanB in the case of vancomycin and other genes that provide resistance and/or tolerance to biocides [29,30]. It should be noted that the gene tetO was not detected in the strains maintained for a while in ceparium. The latter also occurred in the detection of the tetA gene [31,32].
The idea of loss or non-detection of a resistance gene is not new, as this phenomenon has been described in the case of the mecA gene in methicillin-resistant staphylococcal isolates that have remained in the ceparium for some time, and that were then used as control. positive in the PCR [33]. When considering this phenomenon, it is interesting to have more than one native positive control strain, although there is a positive control strain of human origin - which could also be a target of this phenomenon - since it increases the possibility of having at least one of them when It is necessary to carry out the PCR, hence the purpose of this work was to find the less an alternative bacterial strain that can be used as a positive control for the detection of this gene in future research [34-36].
References
- OMS (Organización Mundial De La Salud) (2003) Epidemiología de las infecciones nosocomiales. Cap. 1. In: Prevención de las enfermedades nosocomiales: guía práctica.
- Revelas A (2012) Healthcare-associated infections: A public health problem. Niger Med J 53(2): 59-64.
- David MZ, Daum RS (2010) Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23(3): 616-687.
- Wang L, Ruan S (2017) Modeling Nosocomial Infections of Methicillin- Resistant Staphylococcus aureus with Environment Contamination. Sci Rep 7(1): 580.
- Johnson J (2002) Nosocomial infections. Vet Clin Small Animal 32: 1101- 126.
- Li B, Webster TJ (2018) Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J Orthop Res 36(1): 22-32.
- Garza U, Silva J, Martínez E (2009) Genetics and genomics focused on the study of bacterial resistance. Salud Publica Mex 5: 439-446.
- Peterson E, Kaur P (2018) Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front Microbiol 9: 2928.
- González G, Mella S, Zemelman R, Bello H, Domínguez M (2004) Integrones and genetic resistance cassettes: structure and role against antibacterials. Rev Méd Chile 132(5): 619-626.
- Cabrera C, Gómez R, Zúñiga A (2007) The resistance of bacteria to antibiotics, antiseptics and disinfectants a manifestation of the mechanisms of survival and adaptation. Medical Colombia 38(2): 149-158.
- Domingues S, da Silva GJ, Nielsen KM (2012) Integrons: Vehicles and pathways for horizontal dissemination in bacteria. Mob Genet Elements 2(5): 211-223.
- Martín T (2002) Antimicrobianos que actúan en la pared bacteriana. Botana L, et al. [Eds]. McGraw-Hill Interamericana. Madrid, España. pp 455-467.
- Kong KF, Schneper L, Mathee K (2010) Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS 118(1): 1-36.
- Mediavilla A, García Lobo J (2004) Antibióticos β-lactámicos. Flórez, J, et al. [Eds]. España. pp. 1105-1128.
- Sauvage E, Terrak M (2016) Glycosyltransferases and Transpeptidases/ Penicillin-Binding Proteins: Valuable Targets for New Antibacterials. Antibiotics (Basel) 5(1): E12.
- Chambers HF (1997) Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev 10(4): 781-791.
- Cheesman MJ, Ilanko A, Blonk B, Cock IE (2017) Developing New Antimicrobial Therapies: Are Synergistic Combinations of Plant Extracts/ Compounds with Conventional Antibiotics the Solution? Pharmacogn Rev 11(22): 57-72.
- Weese JS (2005) Methicillin-resistant Staphylococcus aureus: an emerging pathogen in small animals. J Am Anim Hosp Assoc 41(3): 150-157.
- Notario R, Lejona S, Méndez E, All L, Lascialandare S (2007) Insulation of Resisting Staphylococcus Aureus Meticilino Acquired in the Community (Samr-Ac), In Rosario And Santa Fe. Rev Méd Rosario 73: 82-85.
- David, MZ, Daum RS (2010) Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23(3): 616-687.
- Guardabassi L, Loeber M, Jacobson A (2004) Transmission of multiple antimicrobial-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet Microbiol 98(1): 23-27.
- Malik S, Peng H, Barton M (2006) Partial nucleotide sequencing of the mecA genes of Staphylococcus aureus isolates from cats and dogs. J Clin Microbiol 44(2): 413-416.
- Peacock SJ, Paterson GK (2015) Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu Rev Biochem 84: 577-601.
- Guardabassi L, Schwarz S, Lloyd D (2004) Pet animals as reservoirs of antimicrobial-resistant bacteria. J Antimicrob Chemother 54(2): 321- 332.
- Bierowiec K, Płoneczka Janeczko K, Rypuła K (2016) Is the Colonisation of Staphylococcus aureus in Pets Associated with Their Close Contact with Owners? PLoS One 11(5): e0156052.
- Avendaño Rojas, Paulina Alejandra (2010) Identification and study of antimicrobial sensitivity of isolated nosocomial bacteria in veterinary hospital enclosures of the University of Chile. Veterinary Medical Title Report. Fac. Veterinary and Animal Sciences p. 54.
- Wichelhaus T, Kern S, Schäfer V, Brade V (1999) Rapid Detection of Epidemic Strains of Methicillin-Resistant Staphylococcus aureus. J Clin Microbiol 37(3): 690-693.
- Bemis DA, Jones RD, Frank LA, Kania SA (2009) Evaluation of susceptibility test breakpoints used to predict mecA-mediated resistance in Staphylococcus pseudointermedius isolated from dogs. J Vet Diagn Invest 21(1): 53-58.
- Céspedes P, Jara MA, Navarro C (2018) Molecular detection of resistance genes to biocides in Nosocomial Bacterias. EMS Veterinary J 1(1): 006.
- Yáñez D, Jara MA, Navarro C (2018) Detection of blaTEM resistance genes in bacteria described as nosocomial. Global J Sci Frontier Res: D Agriculture and Veterinary 18.
- Galarce N (2011) Detección de tres genes de resistencia a antimicrobianos en cepas de Staphylococcus coagulasa positivas aisladas desde gatos. Memoria Título Médico Veterinario. Fac. Ciencias Veterinarias y Pecuarias.
- Obreque B, Jara MA, Raggi LA, Navarro C (2019) Positive Controls in detection of genes of resistance to tetracyclines in bacteria of veterinary interest. Journal of Veterinary Science and Animal Welfare 3(1): 13-18.
- van Griethuysen A, van Loo I, van Belkum A, Vandenbroucke-Grauls C, Wannet W, et al. (2005) Loss of the mecA gene during storage of methicillin- resistant Staphylococcus aureus strains. J Clin Microbiol 43(3): 1361-1365.
- Graham D, Bergeron G, Bourassa M, Dickson J, Gomes F, et al. (2019) Resistance from Food Animal Production REVIEW Complexities in understanding antimicrobial resistance across domesticated animal, human, and environmental systems. NY Acad Sci 1441: 17-30.
- Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP (2004) Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA 101(26): 9786-9791.
- Said Salim B, Mathema B, Kreiswirth BN (2003) Community-acquired methicillin-resistant Staphylococcus aureus: an emerging pathogen. Infect Control Hosp Epidemiol 24(6): 451-455.






 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.