Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
The Nucleocapsid Protein Gene as Excellent target for Detection of Canine Distemper Virus by Reverse Transcriptase-Polymerase Chain Reaction
*Corresponding author: Navarro C, Department of Preventive Animal Medicine, FAVET, University of Chile, Chile
Received: September 14, 2019;Published: October 01, 2019
DOI: 10.34297/AJBSR.2019.05.000935
Abstract
Canine Distemper is a viral disease of worldwide distribution, lethal and highly contagious, caused by the Canine Distemper Virus, which affects a wide host range, as domestic dogs and wild representatives of different families of carnivores, compromising drastically the conservation of threatened species. For the definitive diagnosis of the ante-mortem disease, have been suggested a variety of clinical parameters and different types of assays, however, due to unpredictable and variable course the e final diagnosis for some animals remains uncertain. Thus, the objective of this work postulated the detection of the nucleocapsid protein gene of Canine Distemper Virus by Reverse Transcriptase-Polymerase Chain Reaction (RTPCR), as a form of rapid and specific diagnosis for the detection of the virus. The specificity of the method was demonstrated by amplification of the expected fragment at 100% of the positive controls to Canine Distemper Virus, both three vaccine controls (Onderstepoort, Lederle and Snyder Hill strains) as ten controls of viral RNA from national isolated, and in the not amplification of the expected fragment in the negative controls (uninfected dogs with and without vaccination). In addition to this, the amplified DNA fragments were sent to sequence and by means the BLAST program, it was confirmed that these were corresponding to Canine Distemper Virus. Additionally, it is proposed that the method could have high sensitivity, due to the amplification of the expected fragment at 91% of the field samples of dogs suspected of Canine Distemper. Based on the above, the method implemented can contribute to the prevention and control of the increase of Canine Distemper, both in the canine population, as in other animals susceptible to the disease.
Keywords: Canine Distemper Virus; Nucleocapsid Protein Gene; RT-PCR
Introduction
The Canine Distemper Virus (CDV) belongs to the genus Morbillivirus of the Paramixoviridae family, Mononegavirales order and is a pleomorphic virus with a lipid envelope, a diameter of 150 to 300 nm, whose genome is a single-stranded RNA, with a negative sense and with a helical nucleocapsid.
The genome consists of about 15.7 kilobases (kb), which codes for six structural proteins: the nucleocapsid protein (N), the phosphoprotein (P), the matrix protein (M), the polymerase (L) and the envelope glycoproteins, the fusion protein (F) and the haemagglutinin (H). These last two, located in the lipid envelope surrounding the virion, are responsible for the recognition and entry of the virus into the host cell [1-4]. The helical nucleocapsid contains the N, P and L proteins, which initiate intracellular replication. The M protein connects the glycoproteins of the surface and nucleocapsid during viral maturation [1,5].
The N gene, of around 1.5 kb, is one of the most conserved genes of the CDV genome and it has highly conserved regions in the first 2/3 of the gene between members of the genus Morbillivirus and encodes the most abundant of the structural viral proteins, the nucleocapsid protein, which has regulatory functions of transcription and replication, as well as the encapsidation of the RNA genome in a resistant RNAsa nucleocapsid. However, despite being a highly conserved region of the CDV genome, some variations of the N gene have been shown among field isolates [6-9].
Diagnosis of CDV. Of the diagnostic methods, the clinical signology is the main tool used by veterinarians, however, the non-specificity of the signs in some cases makes it impossible to reach a final diagnosis, so it is necessary to use laboratory tests. Among them, one of the most used is the ELISA (Enzyme-Linked Immune Sorbent Assay), which can detect serum antibodies (Abs) IgM (against the nucleocapsid proteins, N and P of the CDV) or IgG (against the antigens of the envelope, H and F of the CDV). However, this test does not differentiate if the Abs produced correspond to maternal Abs, vaccine or infection. Otherwise, the production of Abs depends on the stage of the disease and may not be in the initial and final phases of it. In dogs not vaccinated with acute infection, they can die without presenting Abs against the virus. In animals with neurological signs, the determination of specific Abs against CDV in cerebrospinal fluid not contaminated with blood, is a definitive diagnosis of DC encephalitis, however this is a very invasive method and its results are variable [7,10].
Another diagnostic method used is the detection of eosinophilic intracytoplasmic inclusion bodies by cytology or by immunofluorescence, however, these can only be visualized in certain periods of the disease, generally being absent when clinical signs are generated [10,11]. Immunohistochemistry is used to detect viral antigens and/or inclusion bodies in tissues, with reliable results only when there is a marked viremia, and this technique can be performed exclusively on samples taken postmortem [7,9]. The isolation of the virus has a very high sensitivity and specificity, however it is a laborious and very slow procedure, so it is not routinely used for diagnosis [11]. The RT-PCR consists of an exponential amplification of DNA fragments of the virus, after reverse transcription from RNA to complementary DNA, allowing the presence of the virus RNA to be detected early, being a Unlike the other techniques mentioned, a sensitive, specific and rapid test for the diagnosis of CDV can be positive even when other tests fail to detect the virus [7,10,11]. In the last decades, several methods based on RT-PCR for the diagnosis of CDV have been developed, being the main targets for amplification, genomic regions that present a high degree of conservation among the virus isolates. This is how, as in 1999, Frisk et al. developed a method, based on the amplification by RT-PCR of a conserved 287 bp fragment of the N gene, which was later used by several authors, proving to be a very effective technique for the detection of the virus genome [7,8,12]. The main objective of this title memory is to diagnose CDV molecularly, through the implementation of the RT-PCR assay of the nucleocapsid protein gene.
Materials-and-Methods
This work was carried out in the Virology and Microbiology laboratories of the Department of Animal Preventive Medicine of the Faculty of Veterinary and Animal Sciences of the University of Chile, with financing from FIV PROJECT 121014019102010
Controls
Positive controls: For positive vaccinal controls, blood was used from the negative control without vaccination, contaminated with live attenuated CDV virus obtained from the vaccines “Nobivac® Puppy DP” (Onderstepoort strain), “Canigen MHA2PPi/L” (Lederle strain) and “Vanguard” Plus 5/CV-L “(Snyder Hill strain). For this, each vaccine was reconstituted with 0.5 mL of 0.9% sodium chloride, and then this mixture was added to 0.5 mL of blood, to later extract the RNA. The ten positive controls from national isolates of the virus corresponded to viral RNA. These isolates came from domestic dogs from Santiago de Chile and were confirmed as positive by a previous RT-PCR based on the hemagglutinin gene [13,14]. Negative controls: Blood was used with anticoagulant of two uninfected animals, of an adult dog without clinical signs of the disease, without a risk and without vaccination history and of an adult dog without clinical signs of the disease and without a risk history, but with a calendar of daily vaccination (vaccinated for the last time ten months ago).
Suspicious samples of DC
Eleven samples of peripheral blood were analyzed with anticoagulant (2 mL), coming from dogs that showed nervous signs compatible with DC and that in some cases presented positive to the ELISA test specific for CDV, with IgM antibody titers ≥1: 80. These samples were collected from veterinary clinics of Santiago de Chile, maintained in refrigeration at 4 °C and processed in less than two weeks. In addition, they were classified according to race, sex, age, vaccination status against CDV and anti-CDV IgM antibody titers (Table 1 )
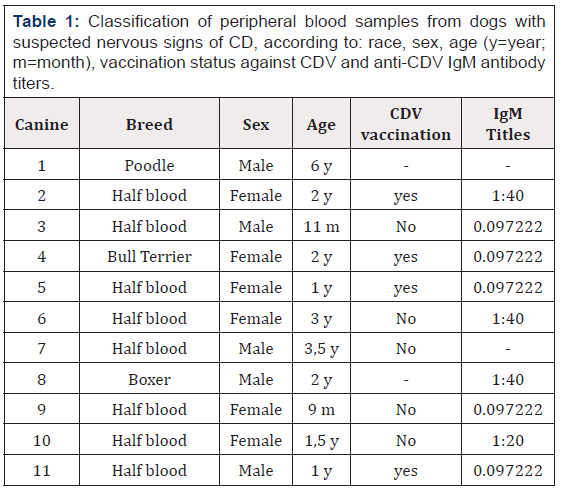
Table 1: Classification of peripheral blood samples from dogs with suspected nervous signs of CD, according to: race, sex, age (y=year; m=month), vaccination status against CDV and anti-CDV IgM antibody titers.
Obtaining the phlogistic layer
In blood samples with anticoagulant, negative controls and animals suspected of DC, the preparation “Histopaque®-1077” from Sigma-Aldrich® was used according to the manufacturer’s instructions, for the isolation of lymphocytes and other mononuclear cells, which are the target of the CDV infection, so a higher concentration of viral RNA was obtained in the samples where the virus was found. For this, 2 mL of Histopaque®-1077 were placed in centrifuge tubes, adding 2 mL of blood with anticoagulant, and then centrifuging at 400xg for 30 minutes at room temperature (RT). The mononuclear band (opaque interface) was then carefully transferred with a Pasteur pipette to a clean centrifuge tube, washed with 10 mL of saline, shaken and centrifuged at 250xg for 10 minutes, to then aspirate. The supernatant and discard it.
Viral RNA extraction using “Trizol LS” kit from Invitrogen®
750 μL of Trizol reagent was added to the mononuclear blood cells, obtained as described above and to 250 μL of the positive vaccine controls (blood contaminated with vaccine), incubating for five minutes at RT. Then, 200 μL of chloroform was added to each tube, they were mixed vigorously for fifteen seconds and incubated at room temperature for five minutes. Then, they were centrifuged at 7000xg for fifteen minutes and the aqueous phase was transferred to a clean tube. For the precipitation of the RNA, 0.5 mL of isopropanol was added, left at room temperature for ten minutes, centrifuged at 7000xg for ten minutes, the supernatant was removed, washed three times with 75% ethanol (1 mL), fifteen were shaken seconds in vortex and centrifuged at 2000xg for five minutes. For the resuspension of the RNA the supernatant was removed, the RNA precipitate was dried under vacuum for five minutes and resuspended in 100 μL of nuclease-free water. Finally, the RNA was incubated at 55-60° C for ten minutes and kept at -20 ° C for later use.
RT-PCR
A 96-well, 96-well Apollo thermocycler (CLP, USA) and a protocol with appropriate temperatures, times and cycles for each stage were used. The primers for the RTPCR were sent to be synthesized to the Bioscan® company, their sequences being in a highly conserved region of the nucleocapsid gene: P1: 5’-ACAGGATTGCTGAGGACCTAT-3’ and P2: 5’-CAAGATAACCATGTACGGTGC-3’. These primers allowed to amplify a fragment of 290 base pairs (bp).
RT-PCR reaction
This was done using the “SuperScript ™ one step RTPCR with platinum Taq” kit (Invitrogen®) according to the manufacturer’s instructions. The reaction began with the synthesis of complementary DNA and a pre-denaturation performed in one cycle (45°C for thirty minutes and then two minutes at 94°C). The PCR amplification was carried out in forty cycles, which consisted of a denaturation (94°C for one minute), alignment of the starters (60°C for two minutes), elongation (72°C for two minutes), and a final extension in one cycle (72°C for ten minutes).
Detection of the DNA fragment synthesized in the RTPCR
The RT-PCR products were analyzed by means of 2% agarose gel electrophoresis in Tris-HC1 buffer (100 mM Tris-HC1, 10 mM EDTA) and comparison against a molecular size standard: Hyperladder IV (Bioline®). The PCR product was mixed with a commercial loading product (Fermentas®) and then the electrophoresis was carried out at 90 V for forty-five minutes. After electrophoresis the gel was incubated with ethidium bromide (0.5 μg/mL) for thirty minutes, then visualized in an ultraviolet light transilluminator and subsequently photographed.
Sequencing of Amplified Fragments
The fragments of DNA obtained from two positive samples were sent to be sequenced, to the Sequencing Center of Genytec Ltda
Bioinformatic analysis of the nucleotide sequences of DNA fragments
The sequences obtained were aligned using the free online program Clustal Ω for obtaining a consensus sequence for each sample used. In addition, the consensus sequences were entered the BLAST program to identify the origin of the DNA fragments obtained in the RT-PCR.
Analysis of results
Those samples that after RT-PCR synthesized a DNA fragment of approximately 290 bp were considered positive and after analysis with the BLAST program their nucleotide identity corresponded to CDV.
Results
Implementation of the RT-PCR assay of the CDV Nucleocapsid Protein Gene
Once the RT-PCR protocol of the nucleocapsid protein gene of the CDV was established, the operation of the method was tested with positive and negative controls, to later analyze the field samples suspected of DC
Controls
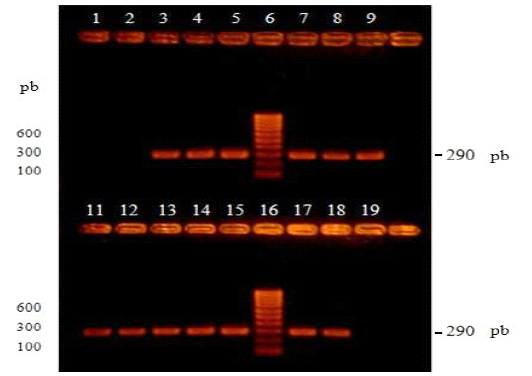
Figure 1: Visualization of products amplified by RT-PCR by electrophoresis in 2% agarose gel and subsequent incubation in ethidium bromide. Lanes 1 and 2 correspond to the negative controls, unvaccinated dog vaccinated and without vaccination respectively. Lanes 3, 4 and 5 correspond to positive vaccinal controls, negative control blood without vaccination contaminated with Onderstepoort, Lederle and Snyder Hill vaccine strains, respectively. Lanes 6 and 16 correspond to the molecular size marker (100-1000 bp). Lanes 7, 8, 9, 11, 12, 13, 14, 15, 17 and 18 correspond to the positive controls of viral RNA from national isolates to CDV.
Three positive controls from commercial vaccines, ten positive controls corresponding to CDV RNA (national isolates) and two negative controls from uninfected dogs were processed. All these controls were subjected to the RT-PCR of the nucleocapsid gene of the CDV and after the visualization of the products, all the positive controls generated intense bands of around 290 bp and in the case of the negative controls, they did not present visible bands (Figure 1 ).
Suspicious samples of DC
Eleven samples of peripheral blood of dogs with nervous signs concordant with DC were collected and, in some cases, they were positive to the specific ELISA test for CDV (IgM) (Table 1 ). All these samples were submitted to the RT-PCR of the nucleocapsid gene of the CDV and after visualization of the products, ten samples (canine 1 to canine 10, Table 1 ) presented bands of around 290 bp, the expected DNA fragment. On the contrary, the sample from canine 11 did not generate visible bands (Figure 2 ).
Controls
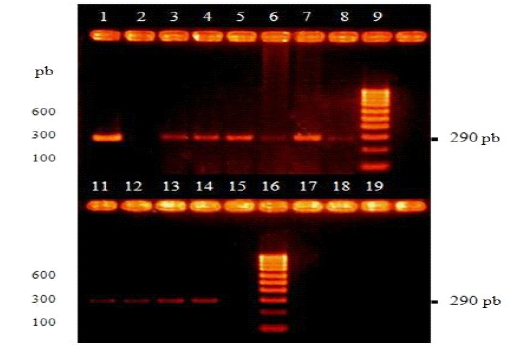
Figure 2: Visualization of amplicons by RT-PCR by electrophoresis in 2% agarose gel and subsequent incubation in ethidium bromide. Lane 1 are the positive vaccine control, Onderstepoort strain. Lane 2 is the negative control, uninfected dog without vaccination. Lane 9 and 16 are the molecular size marker (100-1000 bp). Lanes 3, 4, 5, 6, 7, 8, 11, 12, 13 and 14 are blood samples from canines 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10 respectively. Lane 15 are canine 11 (Table 1 ).
Sequencing of amplified fragments and bioinformatic analysis of the nucleotide sequences of the DNA fragments
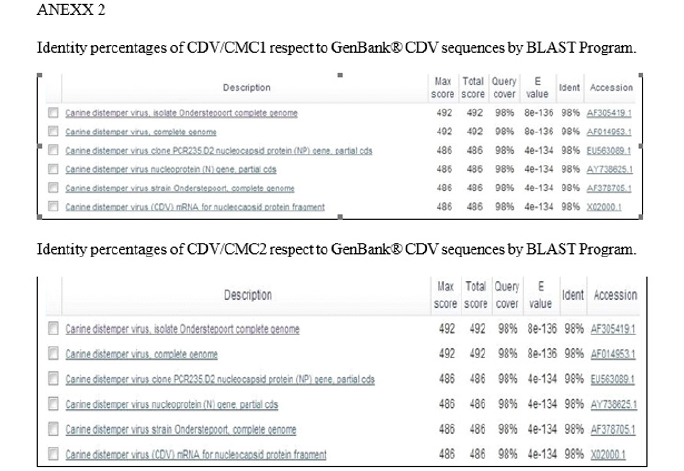
The DNA fragments from two samples considered positive (canine 1 and canine 3, Table 1 ), were sent to be sequenced in triplicate to the Sequencing Center of Genytec Ltd., obtaining five sequences for each sample. The selection criteria for these samples was based on the ages of the animals, where the six-year-old canine 1 was chosen, which is the age where chronic encephalitis would most frequently occur due to CDV infection, which could be related to greater nucleotide variations according to [7]. Likewise, dog 3, eleven months old, was chosen, which is the age in which more animals are usually infected by CDV (less than one year old). The sequences were then aligned using the Clustal Ω program, obtaining a consensus sequence for each sample used: from canine 1 the sequence CDV/CMC1 was obtained and from canine 3 the sequence CDV/CMC2 (Annex 1) was obtained. Next, the consensus sequences were entered into the BLAST program, to identify the origin of the DNA fragments obtained in the RT-PCR, which revealed that the sequence CDV/CMC1 presented a percentage of nucleotide identity of 95% and the CDV/CMC2 sequence of 94%, with respect to the first one hundred results corresponding to CDV, and that in the case of both sequences, these corresponded to CDV (Annex 2).
Discussion
The CD is a disease of worldwide distribution, viral and highly contagious, affecting a wide range of terrestrial carnivores and some marine mammals, which have had a progressive increase in recent years, infecting even animals with their vaccination plans at day. It is due to this increase in hosts, that a true diagnosis of the disease caused by the CDV is crucial, to limit the spread of it to other animals and to give an opportune treatment to the sick. Although the diagnosis of CD is based mainly on the clinical signs of the patient, in certain occasions, it is difficult to reach a final diagnosis in this way, since the main respiratory and digestive signs are common to other diseases that affect the canines and in the case of the more specific clinical signs of DC, such as nervous signs, these most commonly occur in the later stages of the infection, after it has become more widespread. Also, the different clinical presentations of DC vary from one animal to another, making the final diagnosis more complex [1,10]. Although there are several techniques to help the diagnosis of CD, such as virus isolation, immunofluorescence and ELISA, most of these methods are laborious, time-consuming and may give false results, which is why they are convenient for the definitive ante-mortem diagnosis of the disease. For this reason, a sensitive, specific and rapid method is necessary to detect a small amount of the virus early in the infection, which, thanks to the development of molecular biological techniques such as RTPCR, may be possible, where a good white sequence is required. conserved between different strains of the CDV, to avoid mismatches of the splitters and the subsequent failure to amplify it.
In this work a diagnostic method for CDV was implemented through RT-PCR, based on the detection of the nucleocapsid protein gene, a highly conserved region of the CDV genome, which makes this an excellent target gene for the molecular detection of this virus. In fact, several authors have revealed the suitability of molecular biology methods based on the nucleocapsid protein gene for the detection of CDV [1,6-8,12,15]. Additionally, the primers used in this study are in the most conserved region of the nucleocapsid protein gene, the central zone, which would increase the sensitivity of the method [7,8].
Although two Faculty reports based on the CDV RT-PCR have been carried out in our Faculty [13,14], these were based on the hemagglutinin gene, which has the highest antigenic and genetic variation among the CDV genes, presenting about 10% variability between different strains of CDV, which is not the most suitable gene for the diagnosis of the virus [8,11].
In the present study, the previous observations of the RT-PCR utility of the CDV nucleocapsid protein gene were confirmed, with the virus RNA found in about 91% (10/11) of the field samples suspected of DC (Figure 2 ). This result suggests that the protocol used in this title memory, from the obtaining of the phlogistic layer to the visualization of the amplified products, could be highly sensitive. At the same time, this method would be more sensitive than the protocols implemented in previous studies, since [13] only found viral RNA in 7% (3/42) of the samples analyzed that came from dogs suspected of DC and in the case of [13], he found the RNA of the virus in 83% (5/6) of the samples analyzed, which had previously been confirmed as positive by another RT-PCR, however in twenty field samples of dogs suspected of DC, did not find viral RNA [13]. However, to corroborate the sensitivity of the method implemented, additional studies are required. The specificity of the method was corroborated with the amplification of the expected fragment in 100% (10/10) of the positive controls and in the nonamplification of the fragment expected in the negative controls (Figure 1 ). As for the negative controls, it is important to note that one of the animals used for these controls, had its vaccination schedule up to date and had been vaccinated for the last time ten months ago, which supports the preliminary observations that a previous vaccination does not causes false-positive results [7].
On the other hand, CDV can remain active indefinitely at -70°C, -192°C (liquid nitrogen) or lyophilized and only remains active for about a month at -10°C [16], without However, in the positive controls corresponding to CDV RNA (coming from national isolates), it is important to note that this RNA had been stored for more than a year at -20 °C, which is not the ideal temperature to maintain integrity and not degradation, but despite this, the stability of the virus RNA was not affected. Regarding the type of sample chosen for the field samples, blood was chosen with anticoagulant (ethylenediaminetetraacetic acid “EDTA”), because it is one of the most suitable as a substrate for RT-PCR, since it is an easy sample of obtain, maintain cell morphology for a longer time preventing the viral RNA from being degraded by endogenous RNAses when leaving the cell and also the amount of EDTA present in the tubes to obtain blood, would not inhibit the result of the RT-PCR [17]. However, similar results have also been seen with serum and urine samples, further increasing the sensitivity of detection if more than one is used for the diagnosis of the disease [1,7].
Until now, confirmation of CDV infection in live dogs was not useful, mainly due to the low level of sensitivity of the available methods, such as the ELISA test, where the finding of neutralizing antibodies in general does not correlate with the results of the RT-PCR [7], as in this study, where some animals positive for CDV infection had lower antibody titers than those considered positive (<1:80); which could indicate the non-contributory role of neutralizing antibody titers for the etiological diagnosis of the disease.
The field sample suspected of DC that was negative after being analyzed by the proposed method (canine 11, Table 1 ), could have been due to the absence of viral RNA, since either because the virus was not in blood, it could be in other epithelia of the body or because the sample was poorly preserved or poorly processed.
Although DC disease is less frequent in developed countries due to immunization, in recent years, the virus that produces it has emerged as a significant pathogen of vaccinated animal populations, and in the case of this study, the presence of some animals suffering from DC, which were at least once vaccinated against the virus, which raises the hypothesis of failure in the immunization of the vaccine, which could be due to incorrect vaccination plans or genetic variations of the virus [6], however, in order to respond to these presumptions it is necessary to carry out other studies on the subject.
After obtaining the sequences of two of the samples that amplified a fragment of approximately 290 bp and defining the consensus sequence for each of them (Annex 1), the BLAST program was used, where it was obtained that both sequences presented a percentage of nucleotide identity greater than 94% with respect to the first one hundred results corresponding to CDV and that in both cases the amplified fragments belonged to this virus, corroborating the specificity of the method implemented (Annex 2).
The technique implemented, would allow the early isolation of infected animals and the establishment of an appropriate treatment. In addition, in the case of wild animals, the rapid and specific diagnosis given by this method could help the conservation of protected species, which have been at risk due to the dramatic increase in CDV hosts occurred in recent years.
Conclusion
The implemented system represents a fast and specific antemortem diagnosis method of the Canine Distemper disease, being effective for the detection of the virus. The stability of the RNA of the Canine Distemper Virus was not altered when kept for more than one year at -20 °C.
According to the genomic analysis of the sequences CDV/ CMC1 and CDV/CMC2, both showed greater similarity to the Onderstepoort strain of the Canine Distemper Virus and, on the other hand, both sequences showed greater similarity to the American 1 lineage of the Canine Distemper Virus
Based on the results of this study, the implemented method can collaborate with the prevention and control of the increase in Canine Distemper, since it provides a rapid and specific diagnosis for the identification of the responsible virus, which could help improve health of the dog population, as well as that of other animals that are susceptible to the disease
References
- Shin Y, Cho K, Cho H, Kang S, Kim H, et al. (2004) Comparison of one-step RT-PCR and a nested PCR for the detection of canine Distemper virus in clinical samples. Australian Veterinary Journal 82(1-2): 83-86.
- Sidhu M, Husar W, Cook S, Dowling P, Udem S (1993) Canine Distemper terminal and intergenic non-protein coding nucleotide sequences: completion of the entire CDV genome sequence. Virology 193(1): 66-72.
- Summers B, Appel M (1994) Aspects of canine Distemper virus and measles virus encephalomyelitis. Neuropathology and Applied Neurobiology 20: 525- 534.
- Von Messling V, Zimmer G, Herrler G, Haas L, Cattaneo R (2001) The hemagglutinin of canine Distemper virus determines tropism and cytopathogenicity. Journal of Virology 75(14): 6418-6427.
- Beineke A, Puff C, Seehusen F, Baumgartner W (2009) Pathogenesis and immunopathology of systemic and nervous canine Distemper. Vet Immunol Immunopathol 127(1-2): 1-18.
- Castilho J, Brandão P, Carnieli P, Oliveira R, Macedo C, Peixoto Z, Carrieri M, Kotait I (2007) Molecular analysis of the N gene of canine Distemper virus in dogs in Brazil. Arquivo Brasileiro de Medicina Veterinária e Zootecnia 59: 654-659.
- Frisk A, König M, Moritz A, Baumgärtner W (1999) Detection of canine Distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with Distemper. Journal of Clinical Microbiology 37(11): 3634-3643.
- Gallo M, Remorini P, Periolo O, Iglesias M, Mattion N, Latorre J (2007) Detection by RT-PCR and genetic characterization of canine Distemper virus from vaccinated and non-vaccinated dogs in Argentina. Vet Microbiol 125(3-4): 341-349.
- Keawcharoen J, Theamboonlersc A, Jantaradsameec P, Rungsipipatb A, Poovorawanc Y, et al. (2005) Nucleotide sequence analysis of nucleocapsid protein gene of canine Distemper virus isolates in Thailand. Veterinary Microbiology 105: 137-142.
- Appel M, Summers J (1999) Distemper canino: estado actual. Institute for Animal Health, College of Veterinary Medicine, Cornell University, New York, USA. pp. 5.
- Martella V, Elia G, Buonavoglia C (2008) Canine Distemper virus. Veterinary Clinics of North America Small Animal Practice 38: 787-797.
- Calderón M, Remorini P, Periolo O, Iglesias M, Mattion N, Latorre J (2007) Detection by RT‐PCR and genetic characterization of canine Distemper virus from vaccinated and non‐vaccinated dogs in Argentina. Veterinary Microbiology 125(3-4): 341‐349.
- Jara P, Céspedes PF, Navarro C (2018) Canine Distemper Virus detection based in Hemaglutinine Gene as target in Reverse Transcriptase-Polymerase Chain Reaction. Insights Vet Sci 2: 034-041.
- Salas V, Pizarro J, Navarro C (2018) Phylogenetic analysis of canine distemper virus detected in Chile. International Journal of Current Research 10: 72402-72407
- Gámiz C, Martella V, Ulloa R, Fajardo R, Quijano Ii, et al. (2011) Identification of a new genotype of canine Distemper virus circulating in America. Vet Res Commun 35: 381-390.
- Pérez S, Iturbe R, Saldívar E (1993) Caracterización del virus del Distemper en cultivos celulares, aislado de animales clínicamente enfermos. Veterinaria México 24(1): 15-19.
- Khosravinia H, Ramesha K (2007) Influence of EDTA and magnesium on DNA extraction from blood samples and specificity of polymerase chain reaction. African Journal of Biotechnology 6 (3): 184-187.
- Jara C, Matus P, Moreira R (2007) Distemper Canino en Isla Robinson Crusoe (Archipiélago Juan Fernández, V Región): antecedentes de un brote epidémico. Boletín Veterinario Oficial. SAG, Chile 8: 1-19.
- Moreira R, Stutzin M (2005) Estudio de la mortalidad de zorros en la IV Región. Boletín Veterinario Oficial. SAG Chile 3: 1-8.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.