Review Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Can Next Generation Sequencing Be the Standard of Care for all Patients With Metastatic Non-Squamous Non-Small Cell Lung Cancer
*Corresponding author: Francisco J Martinez, Department of Medicine, University of Arizona, USA.
Received: October 30, 2019; Published: November 06, 2019
DOI: 10.34297/AJBSR.2019.06.001002
Introduction
The genetic alterations of most lung cancers are enormous. There are approximately 20,667 genes in the human genome comprised of over 3 billion base pairs. Smokers with lung adenocarcinoma have, on average, 8-10 mutations per 1 million bases (base pairs), which equates to over 50,000 single nucleotides [1]. The current standard of care in the treatment of stage IV nonsquamous non-small cell lung cancer (NSCLC) is based on tailoring treatment on just a handful of actionable genetic drivers. Specifically, the International Association for the Study of Lung Cancer (IASLC), the College of American Pathologists (CAP), and the Association for Molecular Pathology (AMP) issued a joint evidence-based guideline establishing recommendations for molecular diagnostic testing in lung cancer. In summation, their recommendations were to test all patients with lung adenocarcinoma for EGFR and ALK abnormalities regardless of clinical variables such as smoking history, gender, or ethnicity [2]. These guidelines are dated, and the landscape of actionable targets, as well as the availability and cost of technology, have changed dramatically since the recommendation. In light of the explosion of newly discovered gene targets, advances in sequencing technology, and development of novel pharmaceuticals, it is time to reexamine these guidelines and recommend every tumor be sequenced.
The current standard of care for metastatic NSCLC is dependent on if the tumor has EGFR, ALK, ROS-1, BRAF V600, or PDL-1 gene alterations. If the tumor harbors an activating mutation in EGFR exon 19 or 21, an EGFR tyrosine kinase inhibitor is preferred as the first-line therapy [3]. In tumors harboring a ROS-1 or similarly ALK gene alterations, Crizotinib is preferred first-line therapy [4]. In carcinomas with BRAF V600E mutations, the FDA recently approved the combination of Dabrafenib plus Trametinib [5]. If a tumor is PDL-1 positive with a tumor proportion score of ≥50%, single-agent Pembrolizumab is preferred, but only if the tumor does not contain EGFR, ALK, or ROS1 alterations [6]. In the scenario that a tumor has none of the above actionable biomarkers, standard of care is treatment with carboplatin plus pemetrexed or carboplatin plus paclitaxel with or without bevacizumab [7].
Recently, mutations in the MET exon 14 and have been described as a potential target. The MET oncogene has been shown to be a sufficient cause of carcinoma exclusive of other oncogenic drivers including EGFR, KRAS, and BRAF mutations in addition to being exclusive to ALK ROS-1 and RET gene rearrangements [8]. Uniquely, MET mutations are not related to smoking history [8]. Treatment with MET Tki Crizotinib in MET exon 14 positive tumors have shown very high response rates. A single-arm study showing 34 patients with metastatic NSCLC with a MET mutation had median overall survival of 8.1 months, whereas those treated with a MET inhibitor had mean overall survival of 24.6 months [9]. While more evidence is needed, one should consider using a MET Tki as first or second line in patients with MET exon 14 skipping mutations. Despite 3% of lung adenocarcinomas being driven by MET exon 14 skip mutations, a target with known pharmacologic agents, the current standard of care does not currently require genetic testing for MET at diagnosis [10].
The list of possible drug targets continues (see Table 1), and new targets are emerging at a faster pace than ever before. In addition to identifying genes and proteins that are oncogenic drivers, there is another field of small molecules that are emerging as potential biomarkers. JAK and STK11 mutations are potential predictive biomarkers for resistance to immunotherapy, while KRAS mutations, tumor mutational burden, and PDL-1 status may be markers of increased immunotherapy effectiveness [11]. With the simultaneous rise of advanced genomics and an increase in specialized pharmaceuticals, there has been an explosion of newly designed target therapies. When and if a clinician will use these targeted therapies, however, hinges on profiling the carcinoma genome for both breadth and depth of mutations, this is a task most suited for next genome sequencing (NGS). Therefore, as more molecular and gene targets emerge, is it now time to perform NGS on all patients with NSCLC rather than current standard single molecular target testing? (Table 1).
From a clinical perspective, acting on this widespread menu of new actionable targets is only possible if those targets are pursued. Traditional DNA analysis is done with Sanger sequencing (SS), pyrosequencing, or traditional single-gene PCR assays [24]. The major issue with these technologies is only a few genes can be sequenced per run leading to exorbitant cost when looking at more than handful of genes [25]. Additionally, these technologies fail to incorporate intratumor heterogeneity with low sensitivities in identifying oncogenic drivers present at low frequency [26,27]. NGS is a technology in which the entire genome can be analyzed simultaneously, making it an ideal tool when the list of actionable targets is substantial [28].AdditionallyNGS is characterized by high coverage also called “deep sequencing,” which allows for the characterization and identification of intratumor heterogeneity of low-frequency variants [29].
The first task for a clinician using NGS is with obtaining a sample. Analyzing samples can currently originate from FFPE tumor blocks, a diagnostic biopsy sample, or, more recently a simple blood draw (liquid biopsy). FFPE tumor blocks are common and allow the sample to undergo further pathological staining later. Storage of FFPE is also commonplace making it easier for further testing in the future.
With liquid biopsy, one obstacle is yield of sufficient tumor. Currently, this process yields, on average, one cancer cell in 106- 108 cells, which is insufficient genetic material without undergoing an additional enrichment step [30].
This Enrichment step adds another layer of manageable complexity, which is yet another factor a physician needs to consider when interpreting the results. Important aspects to consider when circulating tumor DNA is used for NGS is both the possibility of excluding possible copy number variations though can catch evolving alterations in tumor heterogeneity that would be missed with a solid tissue biopsies [31,32]. Liquid biopsies do offer several advantages, particularly in late-stage disease, where the amount of ctDNA is sufficient. Benefits include being noninvasive and as cheap to run as tumor-based NGS. Liquid biopsies can also be used as a complementary test in addition to tissue testing especially in detecting low-frequency mutation in clonal evolutions or when tissue-based testing is not possible due to risk of biopsy location [33]. Advances in bioinformatic analysis are now at the point where as long as the sample has adequate coverage, liquid NGS is as sensitive as traditional based PCR in identifying mutations [34].
Additional factors include the availability and access to an NGS platform and the amount of time a clinician must wait for sample processing. While ten years ago, NGS was limited to large, wealthy, private institutions, and large university health systems, NGS technology has since been democratized across the continental US. This has been accomplished as all the major NGS business platforms, including Paradigm, Nantomics, and Foundation, have partnered with UPS and other shipping centers and designed specific and affordable NGS-specific shipment packages. While access has increased, turnaround time is still company dependent with one to 4 weeks being typical for NGS [35].
From a cost perspective, Scluckebier has studied the current standard of care for NGS versus EGFR, ALK, and ROS-1. The current standard is to first test for EGFR mutation and follow up a negative test with a FISH for ALK; if ALK is also negative, the rule is to then look at ROS1. NGS is both more sensitive and more specific, with a 24% increase in positive mutations identified; this comes at an extra cost of $400-800.76 compared to the standard [36] Currently there are numerous patient assistance programs supplied by the major companies themselves that cover the costs. Of note, the cost of NGS has fallen by an order of magnitude in the last decade, and a budget impact analysis led by Rosa A. van Amerongen shows cost continuing to drop in the coming years; NGS is already cheaper than if one were to send more than two genes for individual testing [37].
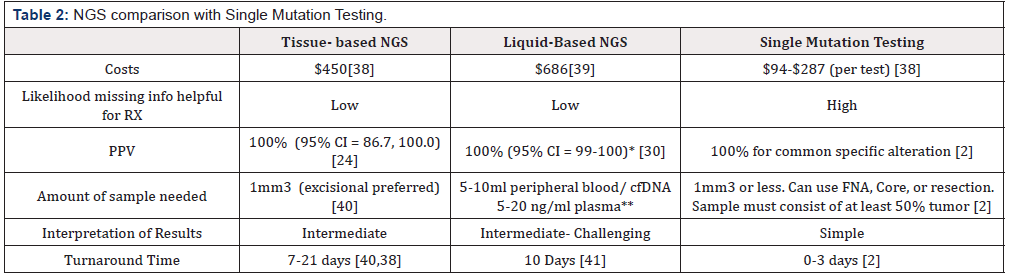
The largest advantage of performing NGS over single mutation testing is the reduction of missing information that would yield an actionable target. Single mutation testing gives just that single gene. When a clinician maps out a course for a patient, having every actionable target at hand is incredibly important. Too much information, however, can present its own set of problems. Often NGS reports are excessively extensive and can be somewhat difficult to interpret due to a large number of mutations and alterations of unknown significance. This presents a catch-22 in data science. One cannot correlate and connect these mutations with clinical outcomes if one does not know they exist; conversely, physicians will not order these tests if there is not a known clinical impact. Be that as it may, the current repertoire of known actionable targets is significant enough to warrant NGS over single mutation testing (Table 2) Depends on gene and stage. Early-stage NSCLC ALK 68.8%, 91.7% ALK 100% EGFR. Fraction of ctDNA is also above 0.4% [42,43] Platform-specific.
Advances in treatment for NSCLC are being driven by an everdeepening and expanding knowledge of the biology of tumor cells on an individual level. Current guidelines are based on the treatment of just a few genes, but one must wonder why not all genes? Every passing year has brought numerous discoveries in gene targets and subsequent successes with novel pharmaceuticals. For instance, this past year saw the identification of pan tumor aberrations, such as NTRK, while extraordinary rare was able to be detected and targeted [44]. The breadth, ease, and cost of testing for these mutations, which can have profound impact on management, needs to be commonplace. Furthermore, making precision genetics part of the standard of care will further drive down costs, and the data generated from NGS will be a boon to knowledge, providing even more insight into other yet undiscovered genes. We believe we are at the point now both from a medical and economic standpoint to recommend that panel testing be routine for patients with NSCLC.
Conflict-of-Interest
None.
References
- Lee W, Zhaoshi Jiang, Jinfeng Liu, Peter M, Haverty, et al. (2010) The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465: 473-477.
- Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, et al. (2013) Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 8(7): 823-859.
- Park K, Tan EH, OByrne K, Zhang L, Boyer M, et al. (2018) Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol 17(5): 577-589.
- Chin LP, Soo RA, Soong R, Ou SH (2012) Targeting ROS1 with anaplastic lymphoma kinase inhibitors: a promising therapeutic strategy for a newly defined molecular subset of non-small-cell lung cancer. J Thorac Oncol 7(11): 1625-1630.
- Planchard D, Besse B, Groen HJM, Souquet PJ, Quoix E, et al. (2017) Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 17(7): 984-993.
- Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, et al. (2016) Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387(10027): 1540-1550.
- Hanna N, Johnson D, Temin S, Baker S Jr, Brahmer J, et al. (2017) Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 35(30): 3484-3515.
- Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, et al. (2015) Activation of MET via Diverse Exon 14 Splicing Alterations Occurs in Multiple Tumor Types and Confers Clinical Sensitivity to MET Inhibitors. Cancer Discov 5(8): 850-859.
- Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, et al. (2017) Impact of MET inhibitors on survival among patients (pts) with MET exon 14 mutant (METdel14) non-small cell lung cancer NSCLC). J Clin Oncol 4(7):721-30.
- Reungwetwattana T, Liang Y, Zhu V, Ou SI (2017) The race to target MET exon 14 skipping alterations in non-small cell lung cancer: The Why, the How, the Who, the Unknown, and the Inevitable. Lung Cancer 103: 27-37.
- Li SD, Ma M, Li H, Waluszko A, Sidorenko T, et al. (2017) Cancer gene profiling in non-small cell lung cancers reveals activating mutations in JAK2 and JAK3 with therapeutic implications. Genome Med 9(1): 89.
- Sharma SV, Bell DW, Settleman J, Haber DA (2007) Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 7: 169-181.
- McKeage K (2015) Alectinib: A Review of Its Use in Advanced ALK-Rearranged Non-Small Cell Lung Cancer. Drugs 75(1): 75-82.
- Camidge DR, Hye Ryun Kim, Myung-Ju Ahn, James Chih-Hsin Yang, Ji-Youn Han, et al. (2018) Brigatinib versus crizotinib in ALK-positive non-small-cell lung cancer. The new England journal of medicine 379: 2027-2039.
- Califano R, Greystoke A, Lal R, Thompson J, Popat S (2017) Management of ceritinib therapy and adverse events in patients with ALK -rearranged non-small cell lung cancer. Lung Cancer 111: 51-58.
- Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, et al. (2016) Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387(10027): 1540-1550.
- Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, et al. (2002) Efficacyand safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 20(3): 719-726.
- Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, et al. (2015) Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 5(8): 842-849.
- Bob TLi, Ronglai Shen, Darren Buonocore, Zachary T Olah, Ai Ni, et al. (2017) Ado-trastuzumab emtansine in patients with HER2 mutant lung cancers: Results from a phase II basket trial. J Clin Oncol 35(24): 2532-2537.
- Drilon A, Rekhtman N, Arcila M, Wang L, Ni A, et al. (2016) Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 17(12): 1653-1660.
- Gautschi O, Zander T, Keller FA, Strobel K, Hirschmann A, et al. (2013) A Patient with Lung Adenocarcinoma and RET Fusion Treated with Vandetanib. J Thorac Oncol 8(5): e43-e44.
- Ou SH, Ahn JS, De Petris L, Govindan R, Yang JC, et al. (2016) Alectinib in Crizotinib-Refractory ALK-Rearranged Non-Small-Cell Lung Cancer: A Phase II Global Study. J Clin Oncol 34(7): 661-668.
- Okimoto RA, Bivona TG (2016) Tracking Down Response and Resistance to TRK Inhibitors. Cancer Discov 6(1): 14-16.
- Vanni I, Coco S, Truini A, Rusmini M, Dal Bello MG, et al. (2015) Next-Generation Sequencing Workflow for NSCLC Critical Samples Using a Targeted Sequencing Approach by Ion Torrent PGMTM Platform. Int J Mol Sci 16(12): 28765-28782.
- Cronin M, Ross JS (2011) Comprehensive next-generation cancer genome sequencing in the era of targeted therapy and personalized oncology. Biomark Med 5(3): 293-305.
- Chen ZY, Zhong WZ, Zhang XC, Su J, Yang X, et al. (2012) EGFR mutation heterogeneity and the mixed response to EGFR tyrosine kinase inhibitors of lung adenocarcinomas. Oncologist 17(7): 978-985.
- Yatabe Y, Matsuo K, Mitsudomi T (2011) Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol 29(22): 2972-2977.
- Gagan J, Van Allen EM (2015) Next-generation sequencing to guide cancer therapy. Genome Med 7(1): 80.
- Meyerson M, Gabriel S, Getz G (2010) Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11(10): 685-696.
- Perakis S, Speicher MR (2017) Emerging concepts in liquid biopsies. BMC Med 15(1): 75.
- Rachiglio AM, Esposito bate R, Sacco A, Pasquale R, Fenizia F, et al. (2016) Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. Oncotarget 7(41): 66595-66605.
- Besse B (2017) Evaluation of liquid biopsies for molecular profiling in untreated patients with stage III/IV non-small cell lung cancer (NSCLC). Journal of Clinical Oncology 35: 11540.
- Villaflor V, Won B, Nagy R, Banks K, Lanman RB, et al. (2016) Biopsy-free circulating tumor DNA assay identifies actionable mutations in lung cancer. Oncotarget 7(41): 66880-66891.
- Köhn L, Johansson M, Grankvist K, Nilsson J (2017) Liquid biopsies in lung cancer—time to implement research technologies in routine care? Ann Transl Med 5(13): 278.
- Yamamoto G, Kikuchi M, Kobayashi S, Arai Y, Fujiyoshi K, et al. (2017) Routine genetic testing of lung cancer specimens derived from surgery, bronchoscopy and fluid aspiration by next generation sequencing. Int J Oncol 50(5): 1579-1589.
- Luciene Schluckebier, Rosangela Caetano, Veronica Aran, CG M F (2017) Cost-effectiveness analysis comparing companion diagnostic tests for EGFR, ALK and ROS-1 versus next-generation sequence (NGS) in advanced adenocarcinoma lung cancer patients. J Clin Oncol 35: 9068-9068.
- Van Amerongen RA, Retèl VP, Coupé VM, Nederlof PM, Vogel MJ, et al. (2016) Next-generation sequencing in NSCLC and melanoma patients: a cost and budget impact analysis. Ecancermedicalscience 10: 684.
- Hamblin A, Wordsworth S, Fermont JM, Page S, Kaur K, et al. (2017) Clinical applicability and cost of a 46-gene panel for genomic analysis of solid tumours: Retrospective validation and prospective audit in the UK National Health Service. PLoSMed 14(2): e1002230.
- Bennett CW, Berchem G, Kim YJ, El-Khoury V (2016) Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget 7(43): 71013-71035.
- Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, et al. (2015) Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 121(4): 631-639.
- Shu Y, Xue Wu, Xiaoling Tong, Xiaonan Wang, Zhili Change, et al. Circulating Tumor DNA Mutation Profiling by Targeted Next Generation Sequencing Provides Guidance for Personalized Treatments in Multiple Cancer Types. Scientific Reports 7: 583.
- Zhang YC, Zhou Q, Wu YL (2017) The emerging roles of NGS-based liquid biopsy in non-small cell lung cancer. Journal of Hematology & Oncology 10(1): 167.
- Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, et al. (2014) An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 20(5): 548-554.
- Amatu A, Sartore-Bianchi A, Siena S (2016) NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 1(2): e000023.




 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.