Mini review 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Epigenetic Markers in Human Diseases
*Corresponding author:Cristiana Libardi Miranda Furtado, Drug Research and Development Center, Postgraduate Program in Medical and Surgical Sciences, Federal University of Ceara, Coronel Nunes de Melo 1127, 60020-181, Fortaleza, Ceara, Brazil.
Received: October 22, 2019; Published: November 12, 2019
DOI: 10.34297/AJBSR.2019.06.001008
Introduction
The human genome project (HGP) provided important information about structure, organization and predicted function of 2.85 billion nucleotides in the DNA sequence, which is the foundation for biomedical research, genetic diagnosis and evolutionary genomics [1]. However, the genetic basis of complex disorders as diabetes, cardiovascular diseases, Alzheimer, schizophrenia, autoimmune diseases and cancer, remains unclear. The multifactorial nature of these diseases involves an intricate gene network and environmental influences that reprogram the genome and hinders the knowledge of the mechanisms involved in its pathophysiology. The environment affects the epigenetic mechanisms that control gene function, without changing the DNA sequence, leading to altered gene expression and disruption of many biological processes. Unlike the genome, the epigenome can be reprogramed and have great plasticity. Epigenetic reprogramming is the main event during cell differentiation, conferring structural and functional differences of a given cell type (phenotype) that is mitotically inheritable ensuring a tissue-specific gene expression.
This phenomenon was first described by Conrad Waddington (1942) in studies of the “Canalization of development”, in which an organism’s genotype responds to the environment in a coordinated manner, in a genotype-genotype and genotype-environment interaction, and this response can be fixed independently of mutational effects [2]. Epigenetic modifications regulate crucial biological process in mammalian development such as genomic imprinting and X-chromosome inactivation (XCI), as well as maintenance of genomic stability and inactivation of repetitive DNA elements [3]. Changes in the establishment or maintenance of epigenetic modifications lead to the development of several human diseases [4,5]. Genomic imprinting, a monoallelic gene expression depending on parental origin, is essential for proper embryo growth and development. Loss of genomic imprinting (LOI) is related to developmental disorders, such as Beckwith-Wiedemann (BWS), Silver-Russell, Prader-Willi and Angelman syndromes [6].
Biallelic expression of imprinted genes has been implicated in the pathogenesis of several complex diseases, such as diabetes mellitus, autism, bipolar disorder, schizophrenia and cancer [7]. The main mechanism that controls the monoallelic expression of imprinted genes is DNA methylation. The imprinting control regions (ICRs) are differentially methylated (DMR) depending on the parental origin [6,7]. DNA methylation is a repressive epigenetic mark characterized by the covalent addition of a methyl group (-CH3) at the carbon-5 position of cytosine-phosphate-guanine dinucleotide (CpG) generating 5-methylcytosine (5mC) (Figure 1A) [3].
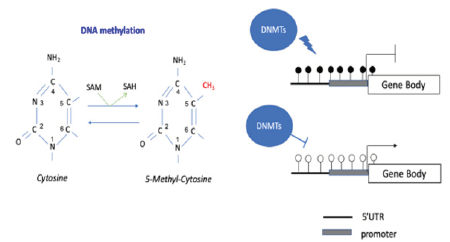
Figure 1A: DNA methyltransferases (DNMTs) adds a methyl group to the cytosine and uses the cofactor S-Adenosylmethionine (SAM), which is converted to S-adenosylhomocysteine (SAH). DNA methylation in the promoter region blocks gene expression or reduces gene activity
In mammals, the DNA methyltransferase (DNMT) enzymes are responsible for the establishment (DNMT 3a e 3b) and maintenance (DNTM1) of a genomic methylation pattern [3]. Demethylation passively occurs during DNA replication and the active demethylation is carried out by the ten–eleven translocation (TET) family that oxidize 5mC to produce 5-hydroxymethylcytosine (5-hmC) [8]. DNA methylation may happen in adenine resides, the N6-methyladenine DNA modification (N6-mA), however, its role in human diseases remain unclear [9]. Usually, mammalian genomes are mostly hypermethylated in non-CpG rich domains (CpG islands), such as promoter and regulatory regions, where the methylation pattern is variable and the hypermethylation at those regions are responsible for gene silencing or reduction of its activity. Hypomethylation of proto-oncogenes and hypermethylation of tumor suppressor genes contribute to cancer development. It has been suggested that aberrant epigenetic reprogramming seems to be the key event that initiates and propagates the carcinogenesis process [10]. DNA methylation is also responsible for the maintenance of genomic stability and silencing of transposable elements [3].
DNA methylation plays an important role in the maintenance of XCI, an epigenetic process by which one of the copies of the X chromosome in the female genome is inactivated. This is a random process with an equal proportion of cells expressing either the maternal or paternal allele, as a dosage compensation mechanism between parental genomes [11]. Skewed XCI is related to X-linked disorders and reproductive alterations, like premature ovarian insufficiency [12]. The main mechanism which controls the XCI is the expression of a long non-coding RNA (lncRNA) called XIST( X-inactive specific transcript) in the inactive X-chromosome. Coding RNAs (RNA) are epigenetic markers that control gene expression and regulate important events during cellular growth and differentiation. The most studied ncRNAs are the microRNAs and lncRNA, a class of transcriptional and pos-transcriptional regulators of gene expression involved in many human diseases, and seems to contribute to the pathogenesis of insulin resistance and glucose dysregulation, neurological and cardiovascular diseases and carcinogenesis [13-15].
The imprinted lncRNA H19 have a controversial role during carcinogenesis, acting as a tumor suppressor or an oncogene, depending on its context within the process of tumor progression [16]. Post-transcriptional modifications at RNA level have been recently reported, a adenosine methylation (N6-methyladenosine, m6A), however its influence in human disease is poorly understood [17]. Post-translational modifications (PTMs) at histone proteins in nucleosomes, change the structure of chromatin and regulate the binding of transcription factors. The most well-recognized histone modifications in humans are acetylation and methylation, however, phosphorylation, ubiquitylation, sumoylation and other modifications can also happen in the histone tail. Specific enzymes are responsible for acetylation, as histone acetyltransferases (HATs) and histone deacetylases (HDACs) (Figure 1B), and methylation, as histone methyltransferases (HMTs) and demethylases (HDMs). Nonetheless, PTMs are a complex event and other enzymes may be involved, especially in the methylation modification [18].
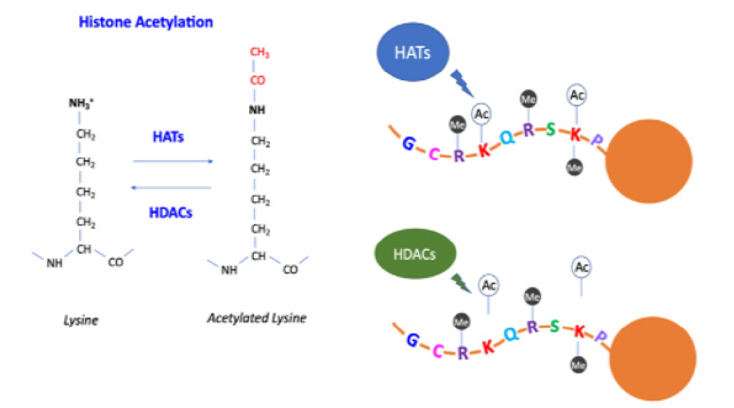
Figure 1B: Histone acetyltransferases (HATs) and histone deacetylases (HDACs) control the acetylation in the amino acids, such as lysine residues at histone tails. Histone acetylation usually leads to transcriptional activation.
Histone modifications have an important role in the maintenance of genomic stability and gene expression regulation, due to the great number of possible modifications, generating a “histone code” that can affect gene expression in different ways. Nonetheless, their functions in human diseases must be better understood. Aberrant reprogramming of epigenetic markers affects embryo development and long-term health in adulthood and the environment and lifestyle can alter the epigenome contributing to the pathogenesis of several diseases in humans. An important information, brought by the HGP to the scientific community, is that the complexity of the human genome is not in the number of protein-coding genes, since less than 2% of the human genome codes for a protein, although a large number of proteins are generated by alternative splicing. Thus, the post-genomic era is focused on the orchestrated transcriptional regulation, function of repetitive elements and pos-translational modifications that can help us understanding the pathogenesis of complex and multifactorial conditions.
Acknowledgement
To the members of the Experimental Oncology Laboratory (LOE) at Drug Research and Development Center (NPDM), Federal University of Ceara. I would like to thank Dr Daniel Pascoalino Pinheiro for the English review.
References
- Collins FS, Lander ES, Rogers J, Waterston RH, Conso IHGS (2004) Finishing the euchromatic sequence of the human genome. Nature 431(7011): 931-945.
- Waddington CH (1942) Canalization of development and the inheritance of acquired characters. Nature 150: 563-565.
- Geiman TM, Muegge K (2010) DNA Methylation in Early Development. Mol Reprod Dev 77(2): 105-113.
- Sharma S, Kelly TK, Jones PA (2010) Epigenetics in cancer. Carcinogenesis 31(1): 27-36.
- Dolinoy DC, Weidman JR, Jirtle RL (2007) Epigenetic gene regulation: Linking early developmental environment to adult disease. Reprod Toxicol 23(3): 297-307.
- Jirtle RL, Weidman JR (2007) Imprinted and more equal. Am Sci 95(2): 143-149.
- Das R, Hampton DD, Jirtle RL (2009) Imprinting evolution and human health. Mamm Genome 20(9-10): 563-572.
- An J, Rao A, Ko M (2017) TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp Mol Med 49(4): e323.
- Xiao CL, Zhu S, He MH, Chen D, Zhang Q, et al. (2018) N-6-Methyladenine DNA Modification in the Human Genome. Mol Cell 71(2): 306-318.
- Feinberg AP, Ohlsson R, Henikoff S (2006) The epigenetic progenitor origin of human cancer. Nat Rev Genet 7(1): 21-33.
- Lyon MF (1962) Sex chromatin and gene action in the mammalian X-chromosome. American journal of human genetics 14: 135-48.
- Miranda-Furtado CL, Luchiari HR, Pedroso DCC, Kogure GS, Caetano LC, et al. (2018) Skewed X-chromosome inactivation and shorter telomeres associate with idiopathic premature ovarian insufficiency. Fertil Steril 110(3): 476-485.
- Satake E, Pezzolesi MG, Dom ZIM, Smiles AM, Niewczas MA, et al. (2018) Circulating miRNA Profiles Associated with Hyperglycemia in Patients with Type 1 Diabetes. Diabetes 67(5): 1013-1023.
- Kim YK, Song J (2018) The Role of Long Noncoding RNAs in Diabetic Alzheimer's Disease. J Clin Med 7(11).
- Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12(12): 861-874.
- Raveh E, Matouk IJ, Gilon M, Hochberg A (2015) The H19 Long non-coding RNA in cancer initiation, progression and metastasis -a proposed unifying theory. Mol Cancer 14: 184.
- Visvanathan A, Somasundaram K (2018) mRNA Traffic Control Reviewed: N6-Methyladenosine (m(6)A) Takes the Driver's Seat. Bioessays 40(1).
- Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21(3): 381-395.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.