Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
First Detection of Emergent Fungal Pathogen Candida auris in Khartoum State, Sudan
*Corresponding author: Ali M Badri, Department of Microbiology, Faculty of Medical Laboratory Sciences, International University of Africa, Khartoum, Sudan.
Received: October 23, 2019; Published: October 29, 2019
DOI: 10.34297/AJBSR.2019.06.000982
Abstract
Background: Candida auris is an emerging multidrug-resistant yeast that can cause invasive infections and is associated with high mortality. Due to high mortality rates and multi-drug-resistant fungus this species is now attracting worldwide attention as Emergent fungal pathogen. The aim of this study to determine the prevalence of Candida auris Among Febrile Patients.
Methods: Blood samples from 100 Febrile Patients were collected and analyzed for the presence of C. auris DNA by PCR.
Results: PCR analysis revealed that 26 out of 100 patients (26%) were positive for C. auris infection.
Conclusion: High level of knowledge and alertness by physicians and healthcare workers, especially in critical care settings, would help to control the spread and improve diagnostic and therapeutic strategies.
Keywords: Candida auris; Candidemia; Febrile Patients; PCR; Khartoum; Sudan
Introduction
Fungal infections represent an important cause of human morbidity and mortality, C. auris is an emerging fungal pathogen that has attracted considerable attention because of its ability to cause infections that are difficult to diagnose and treat [1]. It was first described in 2009 after being isolated from the external ear discharge of a patient in Japan [2]. Outbreaks of C. auris infections have also been reported in patients from United States [3,4], United Kingdom [5,6], South Korea [7,8], South Africa [9,10], India [11,12], Pakistan [9], Israel [13], Venezuela [9,14], Spain [15], Colombia [16], and Kuwait [17]. C. auris is primarily detected in patients with a long period of hospitalization in intensive care units (ICU). C. auris has now been recognized as an important nosocomial pathogen in many countries causing chronic otitis media, bloodstream infections, ventriculitis, osteomyelitis, otomastoiditis, intra-abdominal infections, pleural effusion vulvovaginitis and pericarditis [18,19]. C. auris is resistant to fluconazole (FLU) and is also reported as MICs to all three major antifungal classes [7,20-23] so it may be difficult to start an adequate empirical therapy without accurate species identification. C. auris is usually misidentified by routinely used phenotypic methods in clinical microbiology laboratories. Biochemical assays as one of the most widely used phenotypic approaches, besides of being time consuming and expensive, cannot properly identify [24]. The development of specific PCR assays for C. auris and for C. auris-related species using cultured colonies seems promising for its rapid and accurate identification, particularly in outbreak settings [25,26]. Molecular identification of C. auris can be performed by sequencing various genetic loci, but it is not routinely used. This study was aimed to determine the prevalence of C. auris using PCR in Febrile Patients in Khartoum State, Sudan.
Materials-and-Methods
This was a health facility based descriptive Cross-sectional study. conducted in Khartoum State during the period from January to April 2019. A total of 100 blood samples were collected from febrile patients including 60 males and 40 females with ages ranged from 5 years to 65 years old. complained of fever and other symptoms like headache and general body pain, patient’s positive for Malaria and Typhoid fever were excluded from the study. A structured questionnaire and referring to the patient clinical sheet were being used; demographic data and other Data (Symptoms, using of drug, residence, etc..). From each patient, 3 ml of venous blood sample was collected in sterile EDTA blood containers and frozen at -20ºC until used for DNA extraction. Ethical approval of the study in accordance with the guidelines of the ethical considerations and Patients consent was obtained before the commencement of the study.
DNA Extraction
DNA was extracted by using commercial DNA extraction kit (Intron, Korea) according to the manufacturers’ instructions and stored at -20ºC until used.
Polymerase Chain Reaction (PCR)
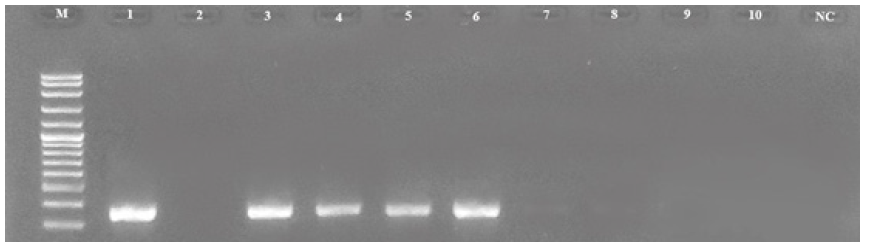
PCR was performed and the test was carried out using following primers: forward: 5’- CGCACATTGCGCCTTGGGGTA-’3and reverse: 5-’GTAGTCCTACCTGATTTGA GGCGAC-’3. Each reaction was performed in total volume of 25 μl, containing 5 μl master mix (Solis Bio dyne master mix), 2 μl of primer, 5 μl of DNA and 13 μl of distilled water. Reactions were performed using PCR machine Techne (UK) under the following cycling conditions: 2 min at 95ºC followed by 30 cycles of 30s at 95°C, 30s at 55°C and 1min at 72°C and final step of 7min at 72°C. 5 μl of the PCR product was analyzed using 1.5% Agarose gel electrophoresis and stained with 0.15% Ethidium bromide and the product was visualized using UV gel documentation. positive reaction was confirmed by the present of 163 bp (Figure 1).
Results and Discussion
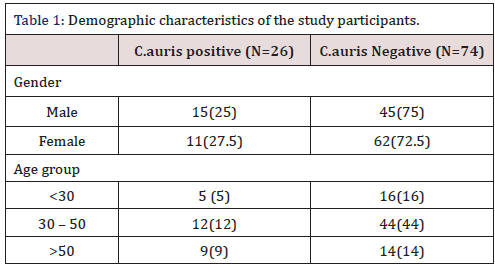
The results of the current study showed prevalence of C. auris- DNA among 26 (26%) febrile patients and negative results among 74 (74%) of the total samples. The results were also clarified according to the gender and age group criteria as the following, according to the gender the C. auris-DNA among male show positive
Candida auris has emerged as a multidrug-resistant yeast causing substantial mortality in health care settings worldwide. Rapid and accurate diagnosis of C. auris infections is crucial to achieve timely and adequate medical treatment and to control fungal outbreaks, Rapid identification of C. auris directly from patient samples is of primary importance for the administration of in 15 (25%) individuals and negative in 45 (75%) individuals, the female show positive in 11(27.5%) individuals and negative in 29 (72.5%) individuals, according to the age group the study individuals was classified into three groups the first group < 30 show positive in 5 (5%) individuals, the second group 30 - 50 show positive results in 12 (12%)individuals, the last group > 50 show positive results in 9 (9%) individuals which clarified in (Table 1).
empirical antifungal therapy. The lack of reliable clinical diagnostic tools potentially led to underestimation and neglection of C. auris cases [27-29]. The ability of C. auris isolates to grow at 40-42˚C, inability to produce hyphae or pseudo hyphae in culture and resistance to fluconazole should prompt clinical microbiology laboratories to seek identification of the yeast isolate by molecular methods [30-33]. The molecular methods provided rapid results within hours of sample processing and highly sensitive compared to the much slower culture results, which may take anywhere from 4 to 14 days and usually misidentified. This is first report regarding C. auris in the Sudan, in the current study the qualitative PCR procedure was used to detect C. auris DNA in blood samples. Specimens were obtained from 100 febrile patients. The results showed positive among 26 febrile patients and negative results among 74 of the total samples. according to the gender the C. auris-DNA among male show positive in 15 (25%) and 11(27.5%) among female. according to the age group the study individuals was classified into three groups the first group < 30 show positive in 5 (5%) individuals, the second group 30 - 50 show positive results in 12 (12%)individuals, the last group > 50 show positive results in 9 (9%) individuals. In most cases, clinical presentation is non-specific and it is often difficult to differentiate between other types of systemic infections. Most of the reported cases in the last 5 years were isolated from blood and other deep-seated sites of infection (including invasive devices and catheters tips). Since the first isolation, C. auris infections have been reported from many countries, including India [11,1], Pakistan [9], South Korea [7,8], Malaysia [34], South Africa [9,10], Oman [35,36], Kenya [37], Kuwait [17], Israel [13], United Arab Emirates [38], Saudi Arabia [39], China [40], Colombia [16], Venezuela [9,14], the United States (US), Spain [15]. The real prevalence and the epidemiology of C. auris still remain uncertain. One of the causes may be the under estimation of its isolation due to the limited accuracy of available conventional diagnostic tools [41].
Conclusion
C. auris is a well-known nosocomial global pathogen that has recently emerged as a significant threat. According to reports of recent outbreaks, colonization is difficult to eradicate, and it tends to persist for months. High level of knowledge and alertness by physicians and healthcare workers, especially in critical care settings, would help to control the spread and improve diagnostic and therapeutic strategies.
Conflict-of-Interest
Authors declare that no conflict of interest exist in this paper
References
- Jeffery Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, et al. (2017) Candida auris: a review of the literature. Clin Microbiol Rev 31(1): e00029-17.
- Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, et al. (2009) Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53(1): 41-44.
- McCarthy M (2016) Hospital transmitted Candida auris infections confirmed in the US. BMJ 355: i5978.
- Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, et al. (2017) Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013-August 2016. Am J Transplant 17(1): 296-299.
- Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, et al. (2016) First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5: 35.
- Borman AM, Szekely A, Johnson EM (2017) Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med Mycol 55(5): 563-567.
- Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, et al. (2011) First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49(9): 3139-3142.
- Kim MN, Shin JH, Sung H, Lee K, Kim EC, et al. (2009) Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 48(6): 57-61.
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, et al. (2017) Simultaneous emergence of multidrugresistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64(2): 134-140.
- Magobo RE, Corcoran C, Seetharam S, Govender NP (2014) Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20(7): 1250-1251.
- Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, et al. (2013) New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19(10):1670-1673.
- Chowdhary A, Anil Kumar V, Sharma C, Prakash A, Agarwal K, et al. (2014) Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33(6): 919-926.
- Ben Ami R, Berman J, Novikov A, Bash E, Shachor Meyouhas Y, et al. (2017) Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis 23(2): 195-203.
- Calvo B, Melo AS, Perozo Mena A, Hernandez M, Francisco EC, et al. (2016) First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect 73(4): 369-374.
- Ruiz Gaitan AC, Moret A, Lopez Hontangas JL, Molina JM, Aleixandre Lopez AI, et al. (2017) Nosocomial fungemia by Candida auris: first four reported cases in continental Europe. Rev Iberoam Micol 34(1): 23-27.
- Morale Lopez SE, Parra Giraldo CM, Ceballos Garzon A, Martinez HP, Rodriguez GJ, et al. (2017) Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis 23(1): 162-164.
- Emara M, Ahmad S, Khan Z, Joseph L, Al ObaidI, Purohit P, et al. (2015) Candida auris candidemia in Kuwait, 2014. Emerg Infect Dis 21(6): 1091-1092.
- Sears D, Schwartz BS (2017) Candida auris: An emerging multidrug-resistant pathogen. Int J Infect Dis 63: 95-98.
- Khan Z, Ahmad S (2017) Candida auris: an emerging multidrug-resistant pathogen of global significance. Curr Med Res Pract 7(6): 240-248.
- Meis JF, Chowdhary A (2018) Candida auris: a global fungal public health threat. Lancet Infect Dis 18(12): 1298-1299.
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, et al. (2017) Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64(2): 134-140.
- Cortegiani A, Misseri G, Fasciana T, Giammanco A, Giarratano A, et al. (2018) Epidemiology, clinical characteristics, resistance and treatment of infections by Candida auris. J Inten Care 6: 69.
- Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, et al. (2018) A multi centre study of antifungal susceptibility patterns among 350 (2009-17) in india: role of the ERG11 and FKS1genes in azole and echinocandin resistance. J Antimicrob Chemother 73(4): 891-899.
- Snayd M, Dias F, Ryan RW, Clout D, Banach DB (2018) Misidentification of Candida auris by Rap ID Yeast Plus, a commercial, biochemical enzyme-based manual rapid identification system. J Clin Microbiol 56(5): 1-2.
- Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, et al. (2017) Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol 55(8): 2445-2452.
- Leach L, Zhu Y, Chaturvedi S (2018) Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J Clin Microbiol 56(2): e01223-17.
- Mizusawa M, Miller H, Green R, et al. (2017). Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55(2): 638-640.
- Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, et al. (2013) New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19(10): 1670-1673.
- Centers for Disease Control and Prevention (2016) Clinical alert to US healthcare facilities-June 2016: global emergence of invasive infections caused by the multidrug-resistant yeast Candida auris. Centers for Disease Control and Prevention, USA.
- Khan Z, Ahmad S (2017) Candida auris: an emerging multidrug-resistant pathogen of global significance. Curr Med Res Pract 7: 240-248.
- CDC (2017) Recommendations for identification of Candida auris.
- Mizusawa M, Miller H, Green R, Lee R, Durante M, et al. (2017) Can a multi-drug resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55(2): 638-640.
- Ben Ami R, Berman J, Novikov A, Bash E, Shachor Meyouhas Y, et al. (2017) Multidrug-resistant Candida haemulonii and C auris. Emerg Infect Dis, Israel 23(1): 195-203.
- Mohd Tap R, Lim TC, Kamarudin NA, Ginsapu SJ, Abd Razak MF, et al. (2018) A fatal case of Candida auris and Candida tropicalis Candidemia in neutropenic patient. Mycopathologia 183(3): 559-564.
- Mohsin J, Hagen F, Al Balushi ZAM, De Hoog GS, Chowdhary A, et al. (2017) The first cases of Candida auris candidaemia in Oman. Mycoses 60(9): 569-575.
- Al Siyabi T, Busaidi Al I, Balkhair A, Al Muharrmi Z, Al Salti M, et al. (2017) First report of Candida auris in Oman: clinical and microbiological description of five candidemia cases. J Inf Secur 75(4): 373-376.
- Okinda N, Kagotho E, Castanheira M, Njuguna A, Omuse G, Makau (2014) P P0065 Candidemia at a referral hospital in sub-Saharan Africa: emergence of Candida auris as a major pathogen. 24th
- Alatoom A, Sartawi M, Lawlor K, AbdelWareth L, Thomsen J, Nusair A, et al. (2018) Persistent candidemia despite appropriate fungal therapy: first case of Candida auris from the United Arab Emirates. Int J Infect Dis 70: 36-37.
- Abdalhamid B, Almaghrabi R, Althawadi S, Omrani A (2018) First report of Candida auris infections from Saudi Arabia. J Infect Public Health 11(4): 598-599.
- Wang X, Bing J, Zheng Q, Zhang F, Liu J, et al. (2018) The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect 7(1): 93.
- Lockhart SR, Berkow EL, Chow N, Welsh RM (2017) Candida auris for the clinical microbiology laboratory: not your grandfather’s Candida species. Clin Microbiol Newsl 39(13): 99-103.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.