Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Covid-19 (SARS-CoV-2) VS Sars-CoV; Summary of all things that healthcare providers should know
*Corresponding author: Seyed Ali Hossein Zahraei, Student research committee at Shiraz university of medical sciences, Shiraz, Iran
Received: April 07, 2020;Published: April 27, 2020
DOI: 10.34297/AJBSR.2020.08.001305
Abstract
The corona viruses are members of a family of enveloped viruses that replicate in the cytoplasm of animal host cells. They are distinguished by the presence of a single-stranded plus-sense RNA genome about 30 kb in length that has a 5′ cap structure and 3′ polyadenylation tract. Upon infection of an appropriate host cell, the 5′-most open reading frame (ORF) of the viral genome is translated into a large polyprotein that is cleaved by viralencoded proteases to release several nonstructural proteins, including an RNA-dependent RNA polymerase (Rep) and an adenosine triphosphatase (ATPase) helicase (Hel). These proteins, in turn, are responsible for replicating the viral genome as well as generating nested transcripts that are used in the synthesis of the viral proteins. The mechanism by which these subgenomic mRNAs are made is not fully understood. The SARS-CoV genomes are about 30 kb in length. Its large RNA genome encodes four structural proteins, sixteen non-structural proteins and eight accessory proteins. SARS-CoV is a plus-strand RNA virus featuring a large single-stranded RNA genome of approximately 29 700 nucleotides. The genome is predicted to consist of atleast fourteen functional ORFs that encode three classes of proteins: two large polyproteins, pp1a and pp1ab, which are cleaved into sixteen non-structural proteins (nsps) required for viral RNA synthesis (and probablyother functions); four structural proteins (the S, E, M and N proteins), essential for viral assembly; and eight accessory proteins, which are thought unimportant in tissue culture but may provide a selective advantage in the infected host.
Introduction
In late 2019, several patients with severe pneumonia were brought into hospitals across the City of Wuhan. The virus causing the pneumonia was sequenced and it was found out that it is a strain of beta-coronavirus and most similarly related to SARS-like BAT coronaviruses bat-SL-CoVZC45 and bat-SL-CoVZXC21 with 88% similarity, 79.5% homology with SARS, and 50% with MERS [1,2]. As a virus transmitted from person to person, as time goes by it inevitably mutates and potentially virulent strains might emerge with high mortality rate [3].
The severe acute respiratory syndrome (SARS) is a novel infectious disease that first occurred in November 2002 in China. SARS is caused by a newly emerged virus belonging to the coronaviridae family, provisionally termed SARS-coronavirus (SARS-CoV) [4]. Available sequence data indicate that SARS-CoV is clearly different from all previously known coronaviruses [5]. Most cases were the result of direct transmission via respiratory droplets during close personal contact, and adequate respiratory protective measures were shown to be effective [6]. In mainland China, around 50% of probable SARS patients did not have an apparent history of close personal contact with another case [7]. At least a proportion of these cases might have arisen from modes of transmission other than droplets.
Methods and Data Gathering
Epidemiology
As of 24 January 2020, 1287 cases of novel coronavirus (2019-nCoV) infections were reported in mainland China, causing 41 deaths. While infections in the first case cluster were initially thought to be mostly due to zoonotic (animal-to-human) transmission—possibly due to wild animals sold at a local seafood wholesale market [8,9] – the growth of case incidence in Wuhan after closure of the market and exportation of cases across China and internationally shows compelling evidence of increasing human-to-human secondary transmission, fueled by human migration. Cases have now been detected in many other parts of the world [10], including other Asian countries, the United States, and France.
The incubation period is defined as the time from infection to illness onset. Knowledge of the incubation period of a directly transmitted infectious disease is critical to determine the time period required for movement restriction of healthy individuals (i.e. quarantine period) [11,12]. A published clinical study has already shown that the average time delay from illness onset to admission is approximately 7 days [13].
Our results show that 95% of incubation periods fall within the range of 2 to 9 days, and the median incubation period was 4-5 days when the Weibull distribution was used as the best-fit model [14]. The median time from illness onset to hospitalization was approximately 3 days. The median time from illness onset to death was 13.8 days, the delay of which is key to appropriate estimation of the case fatality risk for 2019-nCoV [15].
Our estimated median incubation period of 2019-nCoV is comparable to known median values of the incubation period for severe acute respiratory syndrome (SARS)-estimated at 4.0-6.4 days [16-18]. In addition to empirically showing the comparability to SARS, the present study has also shown that the 95th percentile of the incubation period is around 7–8 days, indicating that a nineday quarantine period could mostly ensure the absence of disease among exposed healthy individuals. The time from illness onset to death is also comparable to SARS [14], and the 13.8-day median delay that we calculated indicates that the crude estimation of the ratio of the cumulative number of deaths to that of cases tends to result in underestimation of the case fatality risk, especially during the early stage of the epidemic.
Sars-CoV
Epidemiology: Since November, 2002, an infectious agent has caused outbreaks of an atypical pneumonia in Guangdong Province, southern China [19]. The illness usually began with high fever and mild respiratory symptoms, but rapidly progressed to pneumonia within a few days [20] At the end of February, 2003, the disease had spread to neighbouring regions and countries [21,22]. The disease was severe, transmissible from person to person, and seemed to cause clusters of disease in health-care workers. It was named severe acute respiratory syndrome (SARS), and WHO issued a global alert about the disease on March 13, 2003 [23,24]. SARS outbreaks occurred in South-East Asia, North America, and Europe, and gave rise to the first pandemic of the 21st century. By July 11, 2003, WHO had recorded 8437 cases of SARS worldwide and attibuted 813 deaths to the disease. Most cases were in mainland China and Hong Kong [25]. Although a novel coronavirus, at the end of March, 2003, SARS coronavirus (CoV), was identified as the infectious agent in the syndrome [26-28]. The origin of this pandemic remains unclear. Median values of the incubation period for severe acute respiratory syndrome (SARS) are estimated at 4.0–6.4 days [26].
Result
Genetics
The rapid spread of SARS-CoV-2 raises intriguing questions such as whether its evolution is driven by mutations. To assess the genetic variation, eighty-six complete or near-complete genomes of SARS-CoV-2 were collected from GISAID [https://www.gisaid.org/]. These SARS-CoV-2 strains were detected in infected patients from China (50), USA (11), Australia (5), Japan (5), France (4), Singapore (3), England (2), Taiwan (2), South Korea (1), Belgium (1), Germany (1), and Vietnam (1). The pair-wise nucleotide sequence alignment was performed by ClustalX2, and the sequence of the strain China/ WHU01/2020/EPI_ISL_406716 was used as a reference genome.
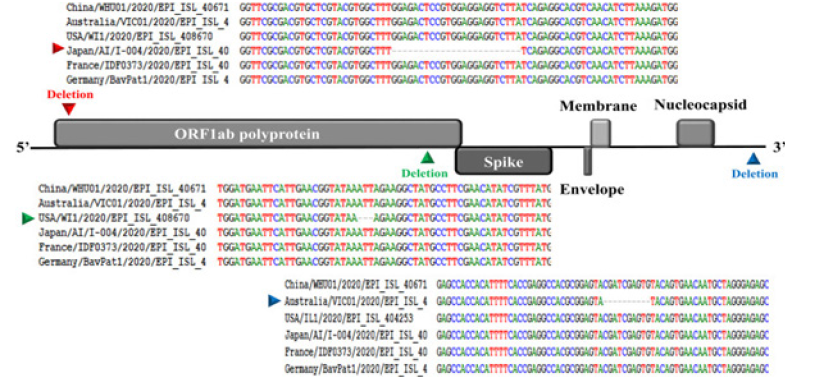
Like other beta corona viruses, the genome of SARS-CoV-2 has a long ORF1ab poly-protein at the 5′ end, followed by four major structural proteins, including the spike surface glycoprotein, small envelope protein, matrix protein, and nucleocapsid protein. Our genetic analysis discovered three deletions in the genomes of SARSCoV- 2 from Japan (Aichi), USA (Wisconsin), and Australia (Victoria) as shown in Figure 1. Two deletions (three nucleotides and twentyfour nucleotides) were in the ORF1ab polyprotein, and one deletion (ten nucleotides) was in the 3′ end of the genome.
Genetics
The corona viruses are members of a family of enveloped viruses that replicate in the cytoplasm of animal host cells [29]. They are distinguished by the presence of a single-stranded plussense RNA genome about 30 kb in length that has a 5′ cap structure and 3′ polyadenylation tract. Upon infection of an appropriate host cell, the 5′-most open reading frame (ORF) of the viral genome is translated into a large polyprotein that is cleaved by viral-encoded proteases to release several nonstructural proteins, including an RNA-dependent RNA polymerase (Rep) and an adenosine triphosphatase (ATPase) helicase (Hel). These proteins, in turn, are responsible for replicating the viral genome as well as generating nested transcripts that are used in the synthesis of the viral proteins. The mechanism by which these subgenomic mRNAs are made is not fully understood. However, recent evidence indicates that transcription-regulating sequences (TRSs) at the 5′ end of each gene represent signals that regulate the discontinuous transcription of subgenomic mRNAs. The TRSs include a partially conserved core sequence (CS) that in some corona viruses is 5′-CUAAAC-3′. Two major models have been proposed to explain the discontinuous transcription in corona viruses and arterio viruses [30,31]. The discovery of transcriptionally active, subgenomic-size minus strands containing the antileader sequence and of transcription intermediates active in the synthesis of mRNAs [32-35] favors the model of discontinuous transcription during the minus strand synthesis [31].
The viral membrane proteins, including the major proteins S (Spike) and M (membrane), are inserted into the endoplasmic reticulum (ER) Golgi intermediate compartment while full-length replicated RNA plus strands assemble with the N (nucleocapsid) protein. This RNA-protein complex then associates with the M protein embedded in the membranes of the ER, and virus particles form as the nucleocapsid complex buds into the lumen of the ER. The virus then migrates through the Golgi complex and eventually exits the cell, likely by exocytosis [29]. The site of viral attachment to the host cell resides within the S protein.
The SARS-CoV genomes are about 30 kb in length [36]. Its large RNA genome encodes four structural proteins, sixteen nonstructural proteins and eight accessory proteins [37]. SARS-CoV is a plus-strand RNA virus featuring a large single-stranded RNA genome of approximately 29, 700 nucleotides [38,39] The genome is predicted to consist of atleast fourteen functional ORFs that encode three classes of proteins: two large poly-proteins, pp1a and pp1ab, which are cleaved into sixteen non-structural proteins(nsps) required for viral RNA synthesis (and probably other functions); four structural proteins (the S, E, M and N proteins), essential for viral assembly; and eight accessory proteins, which are thought unimportant in tissue culture but may provide a selective advantage in theinfected host [40]. The SARS corona virus includes four structural proteins that are required to drive cytoplasmic viral assembly: the spike (S) protein, the membrane (M) protein, the nucleo-capsid (N) protein and the envelope (E) protein [38,39].
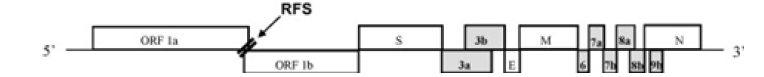
Corona viruses possess the largest genome among any RNA viruses. The genome contains several essential genes that encode the viral proteins necessary for replication, transcription and infectious virus assembly. The essential genes comprise the open reading frames 1a and 1b (ORF1ab) that are translated to produce polyproteins that are involved in viral RNA replication and transcription. In addition to these genes, the other essential genes encode the common viral structural proteins, S, E, M and N, which are involved in infectious virus assembly (Figure 1). Interspersed between these genes in the corona virus genome are several other genes called “group-specific or accessory genes” and their gene products are called “accessory proteins” because many studies using reverse genetics and targeted mutagenesis analysis have shown that these genes and their products are dispensable for virus growth in vitro. Nevertheless, many of these genes are still maintained in the virus genome suggesting that they might play a very important role in the survival of the virus under the natural environment of the infected host.
(Figure 2) The accessory genes are shown as gray boxes. The ORFs 1a and 1b comprise the SARS-CoVreplicase genes. The figure is not drawn to scale. RFS: ribosomal frameshift [41].
Prevention
The wearing of face masks is the single most important precaution against SARS in hospital settings [42] A case-control study among 241 non-infected and 13 infected staff members with documented exposure to 11 index SARS patients suggested that wearing a face mask is the most important infection control tool, followed by appropriate hand hygiene, [43] suggesting that droplets and hands play a major role in transmission of SARSCoV. Hands may be contaminated by patient excretions or contact with contaminated surfaces. From the inanimate environment, nosocomial pathogens can be transmitted to hands quite easily [44]. The World Health Organization (WHO) has suggested that standard disinfectants should be effective against SARS-CoV [45].
Discussion and Conclusion
Incubation
Reports from member laboratories of the WHO SARS network indicated that the virus is stable in faeces and urine at RT for at least 1-2 days and even more stable (up to 4 days) in stool from patients with diarrhoea (which has a higher pH than normal stool) [47,48]; Thermal inactivation of SARS-CoV at 56° and 60°C is highly effective; however, in the presence of protein (20%), infectivity was only reduced by less than 2log10 at 56°C after 30 min (and also after 60 min; data not shown). This has implications for the handling of clinical specimens from SARS patients; for example, pre-treatment of sera at 56°C for 30 min might not be sufficient for full inactivation. On the other hand, as SARS-CoVviraemia does not seem to reach high titres [49], a moderate reduction of residual infectivity might be sufficient to enable the safe performance of serological laboratory assays.
SARS-CoV has been described to persist on surfaces for up to 96 h [50]. In another study, dried SARS-CoV retained its infectivity for as long as six days, indicating a relatively strong survival ability. Only after nine days in a dried state did SARS-CoV completely lose its infectivity [51].
Symptoms
At admission, all patients had fever, sometimes accompanied by myalgia and headache (Table 1). Other symptoms, including dry cough, developed 2–4 days after onset of fever. Shortness of breath (when present) generally manifested in week 2 of illness [52].
As defined by WHO, a suspected case of SARS is an individual with fever (temperature 38°C [100.4°F]), cough or dyspnea, and contact with an individual believed to have SARS or to have traveled to a region where there has been documented transmission of the disease. A probable case is an individual meeting criteria for a suspected case along with radiographic features of pneumonia, respiratory distress syndrome, or an unexplained respiratory illness resulting in death with autopsy results demonstrating pathology of respiratory distress syndrome without an identifiable cause [53].
Treatment
Interferons, a family of cytokines important in the cellular immune response, have been shown to be partly effective against animal and human corona viruses [54-56]. Interferon b was found to be more potent than interferon α or γ, and it remained effective after viral infection, although the potential difference between interferon a and b has been debated [57]. The use of interferons in the treatment of SARS has been limited to interferon a in combination with steroids, immunoglobulins, or thymic peptides, and its efficacy cannot be ascertained [58,59].
A lopinavir–ritonavir co-formulation (Kaletra) is a protease inhibitor preparation used to treat human immunodeficiency virus (HIV) infection. It was used in combination with ribavirin in some Hong Kong hospitals in the hope that it would inhibit coronaviral proteases, thus blocking the processing of the viral replicase polyprotein and preventing the replication of viral RNA. Preliminary results suggest that the use of lopinavir–ritonavir simultaneously with ribavirin and corticosteroids might reduce intubation and mortality rates, especially when administered early [60].
Glycyrrhizin, which is used in the treatment of chronic hepatitis and is relatively nontoxic, has been tested in vitro and found to be an active agent against SARS-CoV [61]. It inhibited viral adsorption and penetration, and was most effective when administered both during and after the viral adsorption period.
Gamma immunoglobulins were used in some hospitals in China and Hong Kong [58]. However, because other therapies such as corticosteroids were often used concomitantly, their effectiveness against SARS remains uncertain.
Noninvasive positive pressure ventilation (NIPPV) was commonly employed in many Chinese hospitals, [58] and was found to avert the need for intubation and invasive ventilation in up to two-thirds of SARS patients with deterioration [62] NIPPV can be given using a continuous positive airway pressure of 4–10cm H2O or bi-level pressure support with an inspiratory positive airway pressure of < 10cm H2O and an expiratory positive airway pressure of 4–6cm H2O. Although NIPPV is useful, the infective risks associated with aerosol generation have hampered its use in many hospitals [63].
References
- Lu R, Zhao X, Li J, Niu P, Yang B, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395 (10224): 565-574.
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, et al. (2020) Complete genome characterisation of a novel coronavirus associated with severe human respiratory disease in Wuhan, China. bioRxiv.
- Wang C, Horby PW, Hayden FG, Gao GF (2020) A novel coronavirus outbreak of global health concern. Lancet 395(10223): 470-473.
- Peng W, Xinxin H, Eric HY, Jessica WY, Kathy L, et al. (2020) Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro Surveillance 25(3): pii 2000044.
- (2020) Center for Disease Control and Prevention. 2019 Novel Coronavirus, Wuhan, China.
- (2020) European Centre for Disease Prevention and Control data. Geographical distribution of 2019- nCov cases.
- Lessler J, Reich NG, Cummings DA (2009) Outbreak of 2009 pandemic influenza A (H1N1) at a New York City school. N Engl J Med 361(27): 2628-2636.
- Nishiura H (2009) Determination of the appropriate quarantine period following smallpox exposure: an objective approach using the incubation period distribution. Int J Hyg Environ Health 212(1): 97-104.
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al. (2020) Clinical Features of Patients Infected With 2019 Novel Coronavirus in Wuhan, Lancet 395(10223): 497-506.
- Lipsitch M, Swerdlow DL, Finelli L (2020) Defining the Epidemiology of Covid-19 - Studies Needed. N Engl J M 382(13): 1194-1196.
- Ghani AC, Donnelly CA, Cox DR, Griffin JT, Fraser C, et al. (2005) Methods for estimating the case fatality ratio for a novel, emerging infectious disease. Am J Epidemiol 162(5): 479-486.
- Donnelly CA, Ghani AC, Leung GM, Hedley AJ, Fraser C, et al. (2003) Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet 361(9371): 1761-1766.
- Cowling BJ, Park M, Fang VJ, Wu P, Leung GM, et al. (2015) Preliminary epidemiological assessment of MERS-CoV outbreak in South Korea, May to June 2015. Euro Surveillance 20(25): 7-13.
- Lessler J, Reich NG, Brookmeyer R, Perl TM, Nelson KE, et al. (2009) Incubation periods of acute respiratory viral infections: a systematic review. Lancet Infect Dis 9(5): 291-300.
- Drosten C, Preiser W, Günther S, Schmitz H, Doerr HW (2003) Severe acute respiratory syndrome:identification of the etiological agent. Trends Mol Med 9(8):325-327.
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, et al. (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300(5624): 1394-1399.
- Seto WH, Tsang D, Yung RW, Ching TY, Ng TK, et al. (2003) Effectiveness of precautions against droplets and contact in prevention of nosocomial transmission of severe acute respiratory syndrome (SARS). Lancet 361(9368): 1519-1520.
- Liang W, Zhu Z, Guo J, Liu Z, He X, et al. (2003) Severe acute respiratory syndrome, Beijing, 2003. Emerg Infect Dis 10(1): 25-31.
- GW Peng, JF He, JY Lin, DH Zhou, DW Yu, et al. (2003) Epidemiological study on severe acute respiratory syndrome in Guangdong province. Chin J Epidemiol 24(5): 350-352.
- (2003) Guangdong Public Health Office. 2003: Document No 2. Summary report of investigating an atypical pneumonia outbreak in Zhongshan.
- KW Tsang, PL Ho, GC Ooi, WK Yee, T Wang, et al. (2003) A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med 348(20): 1977-1985.
- N Lee, D Hui, A Wu, P Chan, P Cameron, et al. (2003) A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 348(20): 1986-1994.
- Update: outbreak of severe acute respiratory syndrome-Worldwide, 2003. MMWR Morb Mort Wkl Rep 52(12): 241-248.
- Severe acute respiratory syndrome (SARS) Wkly Epidemiol Rec 78(12): 81-83.
- Cumulative number of reported probable cases of severe acute respiratory syndrome (SARS) http://www.who.int/csr/sarscountry/2003_07_11/en (accessed July 16, 2003).
- J Peiris, S Lai, L Poon, Y Guan, L Yam, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361(9366): 1319-1325.
- SM Poutanen, DE Low, B Henry, S Finkelstein, D Rose, et al. Identification of severe acute respiratory syndrome in Canada. N Engl J Med 348(20): 1995-2005.
- TG Ksiazek, D Erdman, C Goldsmith, SR Zaki, T Peret, et al. A novel coronavirus associated with severe acute respiratory syndrome N Engl J Med 348(20): 1953-1966.
- BN Fields, DM Knipe, PM Howley, DE Griffin (2001) Fields Virology (Lippincott Williams & Wilkins, Philadelphia, ed. 4, 2001).
- MMC Lai, D Cavanagh (1997) The Molecular Biology of Coronaviruses. Adv Virus Res 48: 1-100.
- SG Sawicki, DL Sawicki (1998) A New Model for Coronavirus Transcription. Adv Exp Med Biol 440: 215-219.
- DL Sawicki et al. (2001) J Gen Virol 82 386.
- SG Sawicki, DL Sawicki (1990) Coronavirus Transcription: Subgenomic Mouse Hepatitis Virus Replicative Intermediates Function in RNA Synthesis. J Virol 64(3): 1050-1056.
- M Schaad, RSJ Baric (1994) J Virol 68 8169.
- PB Sethna et al. (1989) Proc Natl Acad Sci U.S.A. 86 5626.
- Chen LL, Ou HY, Zhang R, Zhang CT (2003) ZCURVE_CoV: a new system to recognize protein coding genes in coronavirus genomes, and its applications in analyzing SARS-CoV genomes. Biochem Biophys Res Commun 307(2): 382-388.
- Bartlam M, Yang H, Rao Z (2005) Structural insights into SARS coronavirus proteins. Curr Opin Struct Biol 15(6): 664-672.
- Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, et al. (2003) The genome sequence of the SARS-associated coronavirus.Science 300(5624): 1399-1404.
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, et al. (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300(5624): 1394-1399.
- Ziebuhr J (2004) Molecular biology of severe acute respiratory syndrome coronavirus. Curr Opin Microbiol 7(4): 412-419.
- Narayanan K, Huang C, Makino S (2008) SARS coronavirus accessory proteins. Virus Res 133(1): 113-121.
- WH Seto, DTsang, RWH Yung, TY Ching, TK Ng, et al. (2003) Effectiveness of precautions against droplets and contact in prevention of nosocomial transmission of severe acute respiratory syndrome (SARS). Lancet 361(9368): 1519-1520.
- (2003) G Kampf Mikrobielle Besiedlung der Hände und ihre epidemiologische Bedeutung. G. Kampf (Ed.), HändehygieneimGesundheitswesen, Springer, Berlin 29-64.
- Seto WH, Tsang D, Yung RW, Ching TY, Ng TK, etal. (2003) Effectiveness of precautions against droplets and contact in prevention of nosocomial transmission of severe acute respiratory syndrome (SARS). Lancet 361(968): 1519-1520.
- DGHMListe der nach den “Richtlinien für die Prüfung chemischer Desinfektionsmittel” geprüften und von der Deutschen Gesellschaft für Hygiene und Mikrobiologie als wirksam befundenen Desinfektionsverfahren. mhp-Verlag, Wiesbaden (2002).
- Jordan LT, Derbyshire JB (1994) Antiviral activity of interferon against transmissible gastroenteritis virus in cell culture and ligated intestinal segments in neonatal pigs. Vet Microbiol 38(3): 263-276.
- Pitkaranta A, Nokso Koivisto J, Jantti V, Takala A, Kilpi T, et al. (1999) Lowered yields of virus-induced interferon production in leukocyte cultures and risk of recurrent respiratory infections in children. J ClinVirol 14(3): 199-205.
- Antonelli G, Scagnolari C, Vicenzi E, Clementi M (2003) Treatment of SARS with human interferons. Lancet 362(9390): 1158-1159.
- Moriguchi H, Sato C (2003) Treatment of SARS with human interferons. Lancet 362(9390): 1159.
- Zhao Z, Zhang F, Xu M, Huang K, Zhong W, et al. (2003) Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J Med Microbiol 52(Pt 8): 715-720.
- Wang J, Liu P, Chen W, Yin S, Jiang S, et al. A hospital outbreak of severe acute respiratory syndrome in Guangzhou, China. Chin Med J (Engl) 116(6): 811-818.
- Sung J (2003) Clinical diagnosis and management of SARS. Proceedings of the WHO Global Conference on Severe Acute Respiratory Syndrome (SARS) 17-18 Kuala Lumpur, Malaysia.
- Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, et al. (2003) Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 361(9374): 2045-2946.
- Xiao Z, Li Y, Chen R, Li S, Zhong S, et al. (2003) A retrospective study of 78 patients with severe acute respiratory syndrome. Chin Med J (Engl) 116(6): 805-810.
- Tsang KW, Lam WK. (2003) Management of severe acute respiratory syndrome: the Hong Kong University experience. Am J Respir Crit Care Med 168(4): 417-424.
- Hsu LY, Lee CC, Green JA, Ang B, Paton NI, et al. (2003) Severe acute respiratory syndrome (SARS) in Singapore: clinical features of index patient and initial contacts. Emerg Infect Dis 9(6): 713-717.
- (2003) World Health Organization. Case definitions for surveillance of severe acute respiratory syndrome (SARS).
- Department of Communicable Disease Surveillance and Response (2003) Consensus document on the epidemiology of severe acute respiratory syndrome (SARS). WHO/CDS/CSR/GAR/2003.11. WHO, Geneva
- (2003) World Health Organization. First data on stability and resistance of SARS coronavirus compiled by members of WHO laboratory.
- Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, et al. (2003) Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361(9371): 1767-1772.
- SM Duan, XS Zhao, RF Wen, JJ Huang, GH Pi, et al. (2003) Stability of SARS coronavirus in human specimens and environment and its sensitivity to heating and UV irradiation. Biomed Environ Sci 16(3): 246-255.
- HF Rabenau, J Cinatl, B Morgenstern, G Bauer, W Preiser, et al. (2005) Stability and inactivation of SARS coronavirus. Med Microbiol Immunol 194(1-2): 1-6.




 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.